Max Westphal
Fraunhofer Institute for Digital Medicine MEVIS, Bremen, Germany
From Data to Diagnosis: A Large, Comprehensive Bone Marrow Dataset and AI Methods for Childhood Leukemia Prediction
Sep 19, 2025Abstract:Leukemia diagnosis primarily relies on manual microscopic analysis of bone marrow morphology supported by additional laboratory parameters, making it complex and time consuming. While artificial intelligence (AI) solutions have been proposed, most utilize private datasets and only cover parts of the diagnostic pipeline. Therefore, we present a large, high-quality, publicly available leukemia bone marrow dataset spanning the entire diagnostic process, from cell detection to diagnosis. Using this dataset, we further propose methods for cell detection, cell classification, and diagnosis prediction. The dataset comprises 246 pediatric patients with diagnostic, clinical and laboratory information, over 40 000 cells with bounding box annotations and more than 28 000 of these with high-quality class labels, making it the most comprehensive dataset publicly available. Evaluation of the AI models yielded an average precision of 0.96 for the cell detection, an area under the curve of 0.98, and an F1-score of 0.61 for the 33-class cell classification, and a mean F1-score of 0.90 for the diagnosis prediction using predicted cell counts. While the proposed approaches demonstrate their usefulness for AI-assisted diagnostics, the dataset will foster further research and development in the field, ultimately contributing to more precise diagnoses and improved patient outcomes.
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022
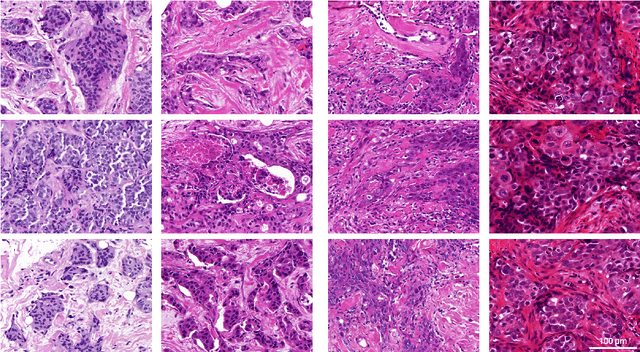
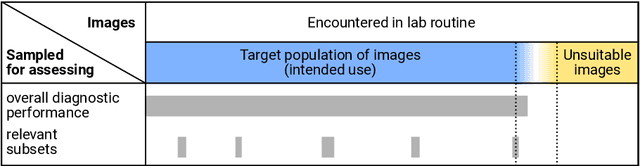
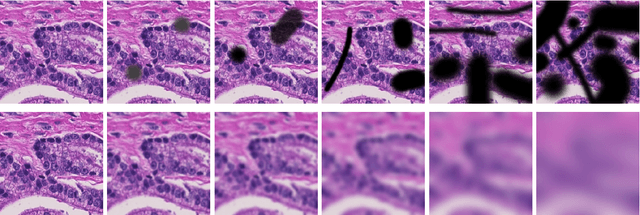
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
A multiple testing framework for diagnostic accuracy studies with co-primary endpoints
Nov 08, 2019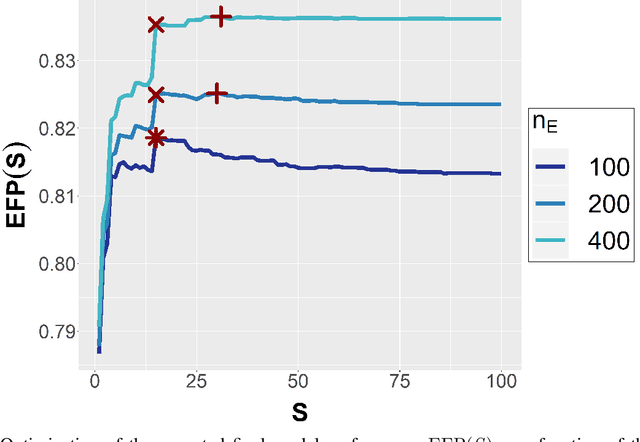
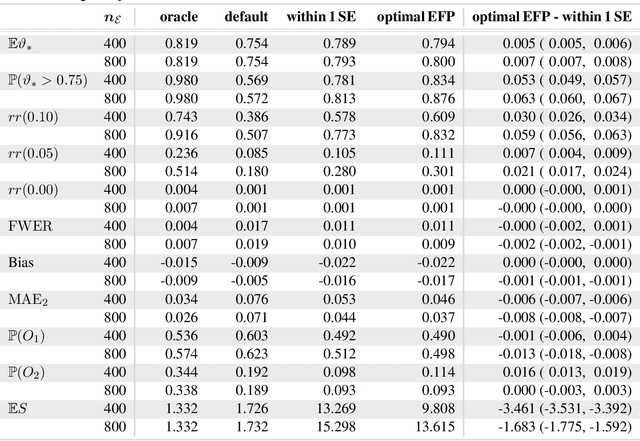
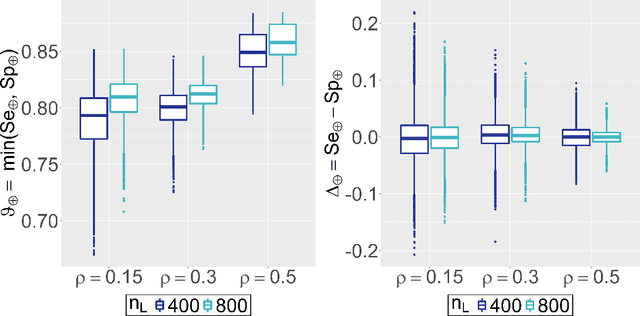
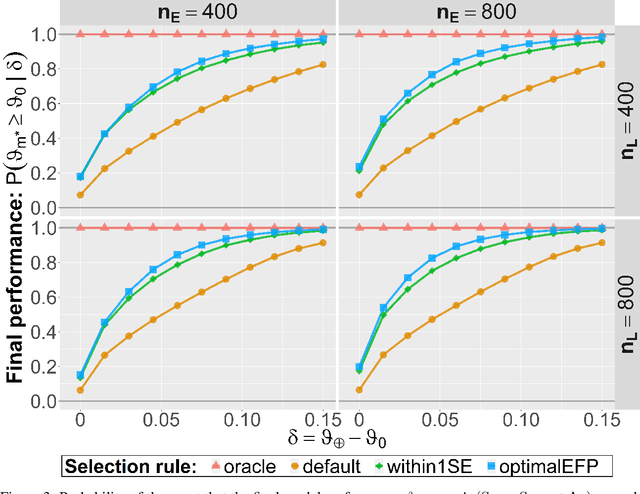
Abstract:Major advances have been made regarding the utilization of artificial intelligence in health care. In particular, deep learning approaches have been successfully applied for automated and assisted disease diagnosis and prognosis based on complex and high-dimensional data. However, despite all justified enthusiasm, overoptimistic assessments of predictive performance are still common. Automated medical testing devices based on machine-learned prediction models should thus undergo a throughout evaluation before being implemented into clinical practice. In this work, we propose a multiple testing framework for (comparative) phase III diagnostic accuracy studies with sensitivity and specificity as co-primary endpoints. Our approach challenges the frequent recommendation to strictly separate model selection and evaluation, i.e. to only assess a single diagnostic model in the evaluation study. We show that our parametric simultaneous test procedure asymptotically allows strong control of the family-wise error rate. Moreover, we demonstrate in extensive simulation studies that our multiple testing strategy on average leads to a better final diagnostic model and increased statistical power. To plan such studies, we propose a Bayesian approach to determine the optimal number of models to evaluate. For this purpose, our algorithm optimizes the expected final model performance given previous (hold-out) data from the model development phase. We conclude that an assessment of multiple promising diagnostic models in the same evaluation study has several advantages when suitable adjustments for multiple comparisons are conducted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge