André Homeyer
Explainable histomorphology-based survival prediction of glioblastoma, IDH-wildtype
Jan 16, 2026Abstract:Glioblastoma, IDH-wildtype (GBM-IDHwt) is the most common malignant brain tumor. Histomorphology is a crucial component of the integrated diagnosis of GBM-IDHwt. Artificial intelligence (AI) methods have shown promise to extract additional prognostic information from histological whole-slide images (WSI) of hematoxylin and eosin-stained glioblastoma tissue. Here, we present an explainable AI-based method to support systematic interpretation of histomorphological features associated with survival. It combines an explainable multiple instance learning (MIL) architecture with a sparse autoencoder (SAE) to relate human-interpretable visual patterns of tissue to survival. The MIL architecture directly identifies prognosis-relevant image tiles and the SAE maps these tiles post-hoc to visual patterns. The MIL method was trained and evaluated using a new real-world dataset that comprised 720 GBM-IDHwt cases from three hospitals and four cancer registries in Germany. The SAE was trained using 1878 WSIs of glioblastoma from five independent public data collections. Despite the many factors influencing survival time, our method showed some ability to discriminate between patients living less than 180 days or more than 360 days solely based on histomorphology (AUC: 0.67; 95% CI: 0.63-0.72). Cox proportional hazards regression confirmed a significant difference in survival time between the predicted groups after adjustment for established prognostic factors (hazard ratio: 1.47; 95% CI: 1.26-1.72). Our method identified multiple interpretable visual patterns associated with survival. Three neuropathologists separately found that 21 of the 24 most strongly associated patterns could be clearly attributed to seven histomorphological categories. Necrosis and hemorrhage appeared to be associated with shorter survival while highly cellular tumor areas were associated with longer survival.
Automatic Extraction of Rules for Generating Synthetic Patient Data From Real-World Population Data Using Glioblastoma as an Example
Dec 18, 2025Abstract:The generation of synthetic data is a promising technology to make medical data available for secondary use in a privacy-compliant manner. A popular method for creating realistic patient data is the rule-based Synthea data generator. Synthea generates data based on rules describing the lifetime of a synthetic patient. These rules typically express the probability of a condition occurring, such as a disease, depending on factors like age. Since they only contain statistical information, rules usually have no specific data protection requirements. However, creating meaningful rules can be a very complex process that requires expert knowledge and realistic sample data. In this paper, we introduce and evaluate an approach to automatically generate Synthea rules based on statistics from tabular data, which we extracted from cancer reports. As an example use case, we created a Synthea module for glioblastoma from a real-world dataset and used it to generate a synthetic dataset. Compared to the original dataset, the synthetic data reproduced known disease courses and mostly retained the statistical properties. Overall, synthetic patient data holds great potential for privacy-preserving research. The data can be used to formulate hypotheses and to develop prototypes, but medical interpretation should consider the specific limitations as with any currently available approach.
Applications of artificial intelligence in the analysis of histopathology images of gliomas: a review
Feb 05, 2024Abstract:In recent years, the diagnosis of gliomas has become increasingly complex. Analysis of glioma histopathology images using artificial intelligence (AI) offers new opportunities to support diagnosis and outcome prediction. To give an overview of the current state of research, this review examines 70 publicly available research studies that have proposed AI-based methods for whole-slide histopathology images of human gliomas, covering the diagnostic tasks of subtyping (16/70), grading (23/70), molecular marker prediction (13/70), and survival prediction (27/70). All studies were reviewed with regard to methodological aspects as well as clinical applicability. It was found that the focus of current research is the assessment of hematoxylin and eosin-stained tissue sections of adult-type diffuse gliomas. The majority of studies (49/70) are based on the publicly available glioblastoma and low-grade glioma datasets from The Cancer Genome Atlas (TCGA) and only a few studies employed other datasets in isolation (10/70) or in addition to the TCGA datasets (11/70). Current approaches mostly rely on convolutional neural networks (53/70) for analyzing tissue at 20x magnification (30/70). A new field of research is the integration of clinical data, omics data, or magnetic resonance imaging (27/70). So far, AI-based methods have achieved promising results, but are not yet used in real clinical settings. Future work should focus on the independent validation of methods on larger, multi-site datasets with high-quality and up-to-date clinical and molecular pathology annotations to demonstrate routine applicability.
Digitization of Pathology Labs: A Review of Lessons Learned
Jun 07, 2023


Abstract:Pathology laboratories are increasingly using digital workflows. This has the potential of increasing lab efficiency, but the digitization process also involves major challenges. Several reports have been published describing the individual experiences of specific laboratories with the digitization process. However, a comprehensive overview of the lessons learned is still lacking. We provide an overview of the lessons learned for different aspects of the digitization process, including digital case management, digital slide reading, and computer-aided slide reading. We also cover metrics used for monitoring performance and pitfalls and corresponding values observed in practice. The overview is intended to help pathologists, IT decision-makers, and administrators to benefit from the experiences of others and to implement the digitization process in an optimal way to make their own laboratory future-proof.
The NCI Imaging Data Commons as a platform for reproducible research in computational pathology
Mar 16, 2023



Abstract:Objective: Reproducibility is critical for translating machine learning-based (ML) solutions in computational pathology (CompPath) into practice. However, an increasing number of studies report difficulties in reproducing ML results. The NCI Imaging Data Commons (IDC) is a public repository of >120 cancer image collections, including >38,000 whole-slide images (WSIs), that is designed to be used with cloud-based ML services. Here, we explore the potential of the IDC to facilitate reproducibility of CompPath research. Materials and Methods: The IDC realizes the FAIR principles: All images are encoded according to the DICOM standard, persistently identified, discoverable via rich metadata, and accessible via open tools. Taking advantage of this, we implemented two experiments in which a representative ML-based method for classifying lung tumor tissue was trained and/or evaluated on different datasets from the IDC. To assess reproducibility, the experiments were run multiple times with independent but identically configured sessions of common ML services. Results: The AUC values of different runs of the same experiment were generally consistent and in the same order of magnitude as a similar, previously published study. However, there were occasional small variations in AUC values of up to 0.044, indicating a practical limit to reproducibility. Discussion and conclusion: By realizing the FAIR principles, the IDC enables other researchers to reuse exactly the same datasets. Cloud-based ML services enable others to run CompPath experiments in an identically configured computing environment without having to own high-performance hardware. The combination of both makes it possible to approach the reproducibility limit.
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022
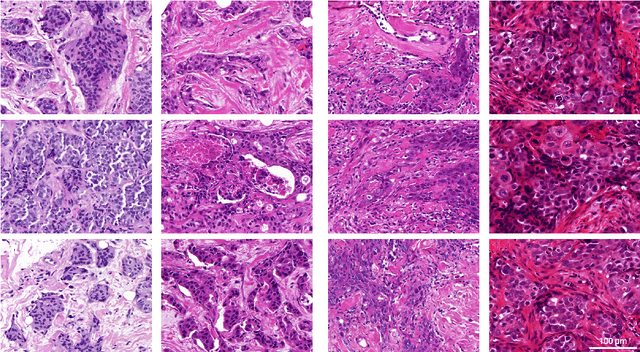
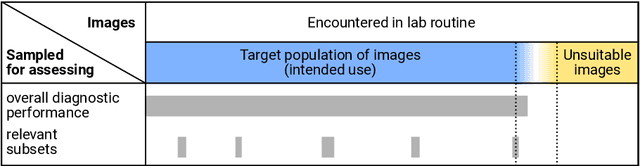
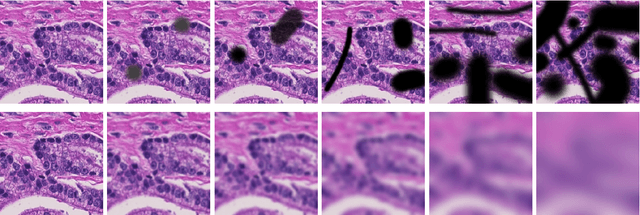
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
Evaluating Generic Auto-ML Tools for Computational Pathology
Dec 07, 2021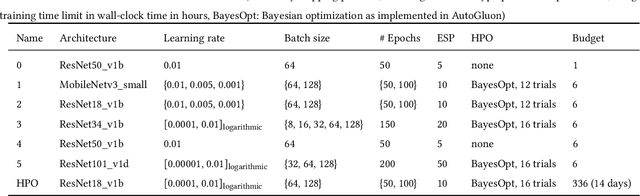
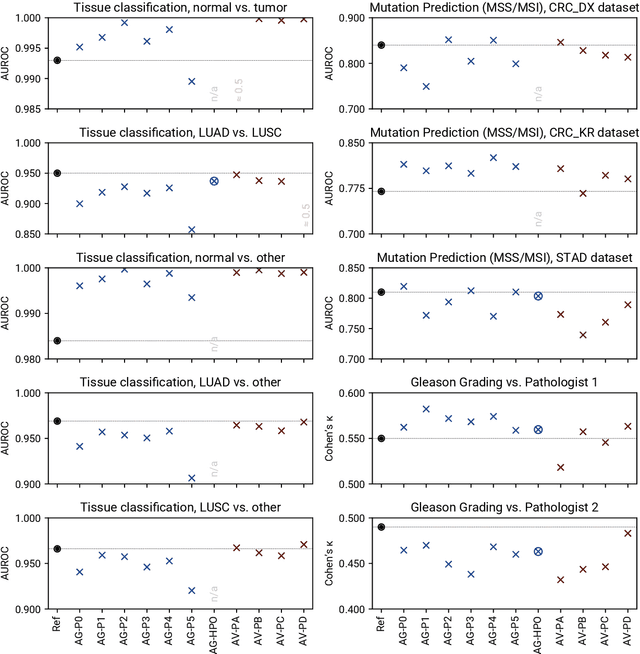

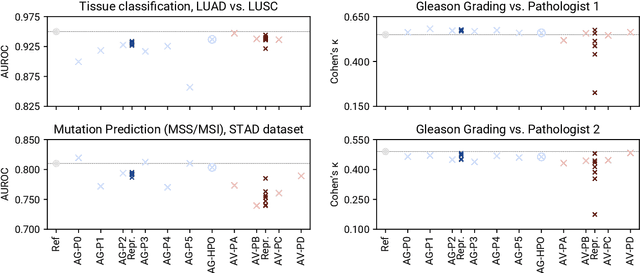
Abstract:Image analysis tasks in computational pathology are commonly solved using convolutional neural networks (CNNs). The selection of a suitable CNN architecture and hyperparameters is usually done through exploratory iterative optimization, which is computationally expensive and requires substantial manual work. The goal of this article is to evaluate how generic tools for neural network architecture search and hyperparameter optimization perform for common use cases in computational pathology. For this purpose, we evaluated one on-premises and one cloud-based tool for three different classification tasks for histological images: tissue classification, mutation prediction, and grading. We found that the default CNN architectures and parameterizations of the evaluated AutoML tools already yielded classification performance on par with the original publications. Hyperparameter optimization for these tasks did not substantially improve performance, despite the additional computational effort. However, performance varied substantially between classifiers obtained from individual AutoML runs due to non-deterministic effects. Generic CNN architectures and AutoML tools could thus be a viable alternative to manually optimizing CNN architectures and parametrizations. This would allow developers of software solutions for computational pathology to focus efforts on harder-to-automate tasks such as data curation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge