Matthew B. A. McDermott
Harvard Medical School
A Comprehensive Evaluation of Multi-task Learning and Multi-task Pre-training on EHR Time-series Data
Jul 20, 2020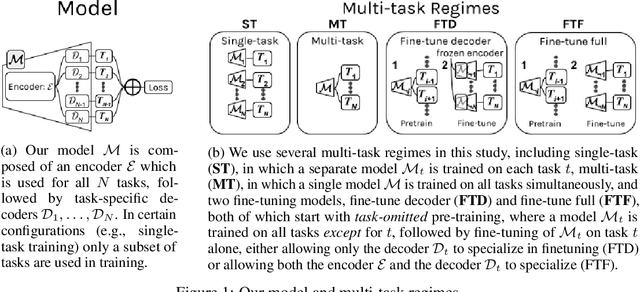
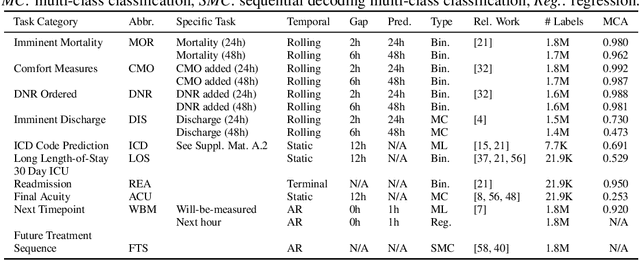
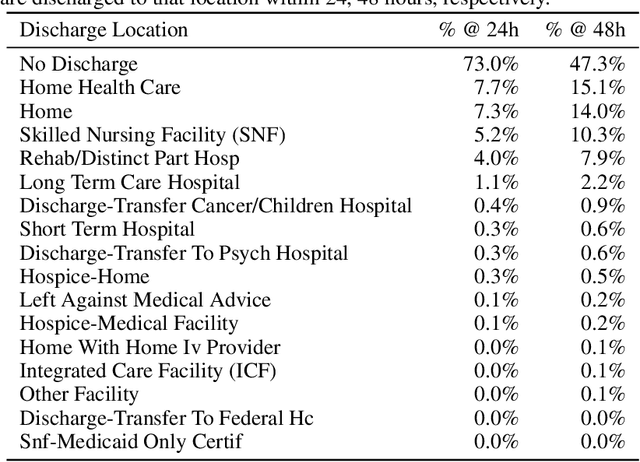
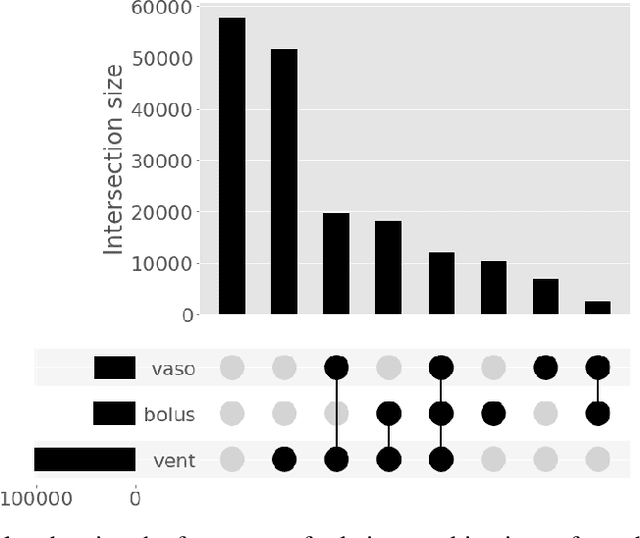
Abstract:Multi-task learning (MTL) is a machine learning technique aiming to improve model performance by leveraging information across many tasks. It has been used extensively on various data modalities, including electronic health record (EHR) data. However, despite significant use on EHR data, there has been little systematic investigation of the utility of MTL across the diverse set of possible tasks and training schemes of interest in healthcare. In this work, we examine MTL across a battery of tasks on EHR time-series data. We find that while MTL does suffer from common negative transfer, we can realize significant gains via MTL pre-training combined with single-task fine-tuning. We demonstrate that these gains can be achieved in a task-independent manner and offer not only minor improvements under traditional learning, but also notable gains in a few-shot learning context, thereby suggesting this could be a scalable vehicle to offer improved performance in important healthcare contexts.
CheXpert++: Approximating the CheXpert labeler for Speed,Differentiability, and Probabilistic Output
Jun 26, 2020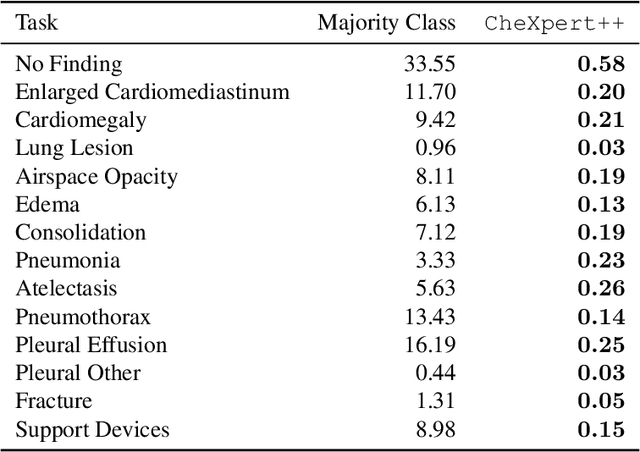
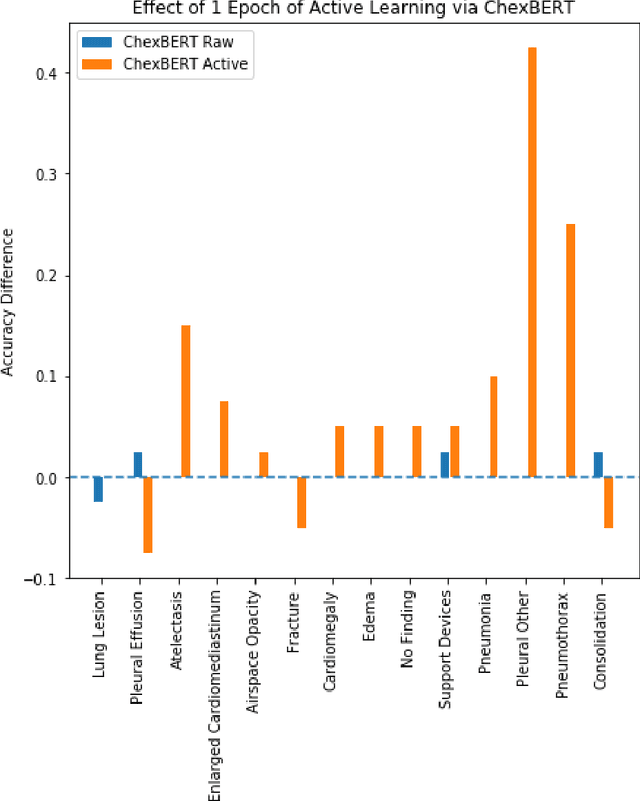
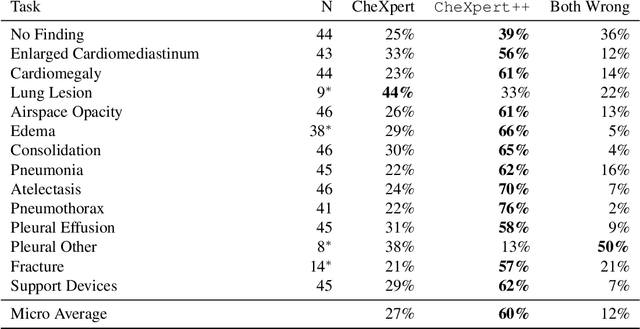
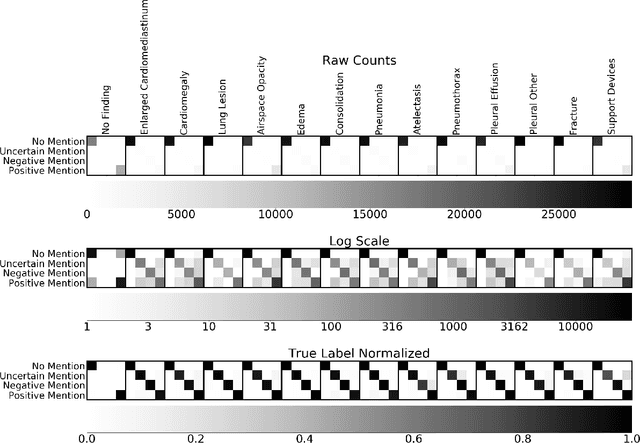
Abstract:It is often infeasible or impossible to obtain ground truth labels for medical data. To circumvent this, one may build rule-based or other expert-knowledge driven labelers to ingest data and yield silver labels absent any ground-truth training data. One popular such labeler is CheXpert, a labeler that produces diagnostic labels for chest X-ray radiology reports. CheXpert is very useful, but is relatively computationally slow, especially when integrated with end-to-end neural pipelines, is non-differentiable so can't be used in any applications that require gradients to flow through the labeler, and does not yield probabilistic outputs, which limits our ability to improve the quality of the silver labeler through techniques such as active learning. In this work, we solve all three of these problems with $\texttt{CheXpert++}$, a BERT-based, high-fidelity approximation to CheXpert. $\texttt{CheXpert++}$ achieves 99.81\% parity with CheXpert, which means it can be reliably used as a drop-in replacement for CheXpert, all while being significantly faster, fully differentiable, and probabilistic in output. Error analysis of $\texttt{CheXpert++}$ also demonstrates that $\texttt{CheXpert++}$ has a tendency to actually correct errors in the CheXpert labels, with $\texttt{CheXpert++}$ labels being more often preferred by a clinician over CheXpert labels (when they disagree) on all but one disease task. To further demonstrate the utility of these advantages in this model, we conduct a proof-of-concept active learning study, demonstrating we can improve accuracy on an expert labeled random subset of report sentences by approximately 8\% over raw, unaltered CheXpert by using one-iteration of active-learning inspired re-training. These findings suggest that simple techniques in co-learning and active learning can yield high-quality labelers under minimal, and controllable human labeling demands.
ML4H Abstract Track 2019
Feb 05, 2020Abstract:A collection of the accepted abstracts for the Machine Learning for Health (ML4H) workshop at NeurIPS 2019. This index is not complete, as some accepted abstracts chose to opt-out of inclusion.
Cross-Language Aphasia Detection using Optimal Transport Domain Adaptation
Dec 04, 2019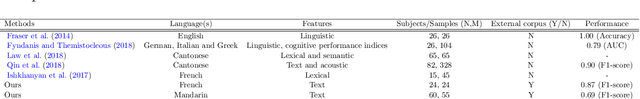
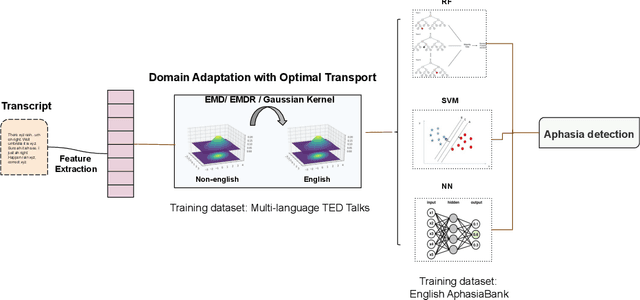
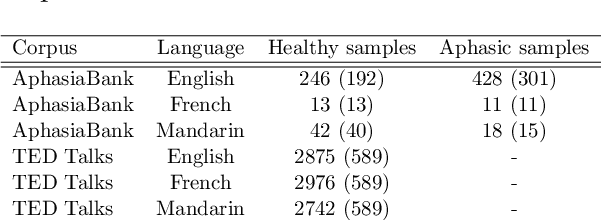
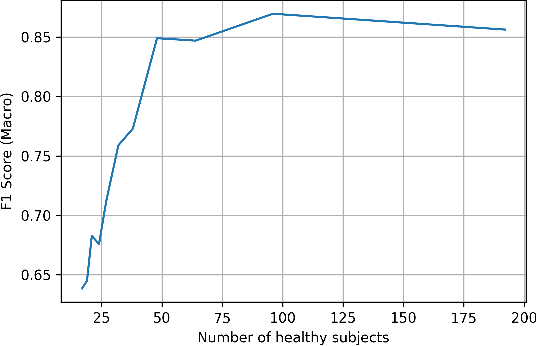
Abstract:Multi-language speech datasets are scarce and often have small sample sizes in the medical domain. Robust transfer of linguistic features across languages could improve rates of early diagnosis and therapy for speakers of low-resource languages when detecting health conditions from speech. We utilize out-of-domain, unpaired, single-speaker, healthy speech data for training multiple Optimal Transport (OT) domain adaptation systems. We learn mappings from other languages to English and detect aphasia from linguistic characteristics of speech, and show that OT domain adaptation improves aphasia detection over unilingual baselines for French (6% increased F1) and Mandarin (5% increased F1). Further, we show that adding aphasic data to the domain adaptation system significantly increases performance for both French and Mandarin, increasing the F1 scores further (10% and 8% increase in F1 scores for French and Mandarin, respectively, over unilingual baselines).
Approaching Small Molecule Prioritization as a Cross-Modal Information Retrieval Task through Coordinated Representation Learning
Nov 22, 2019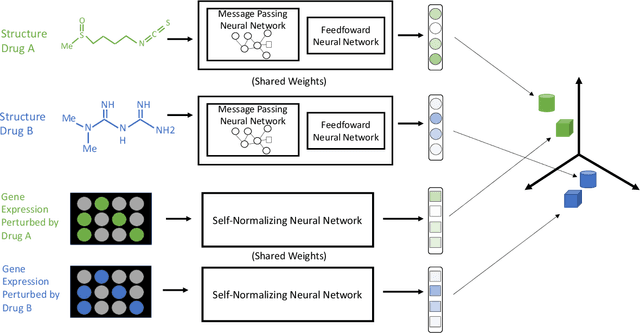
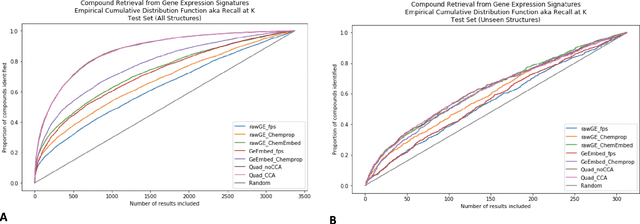
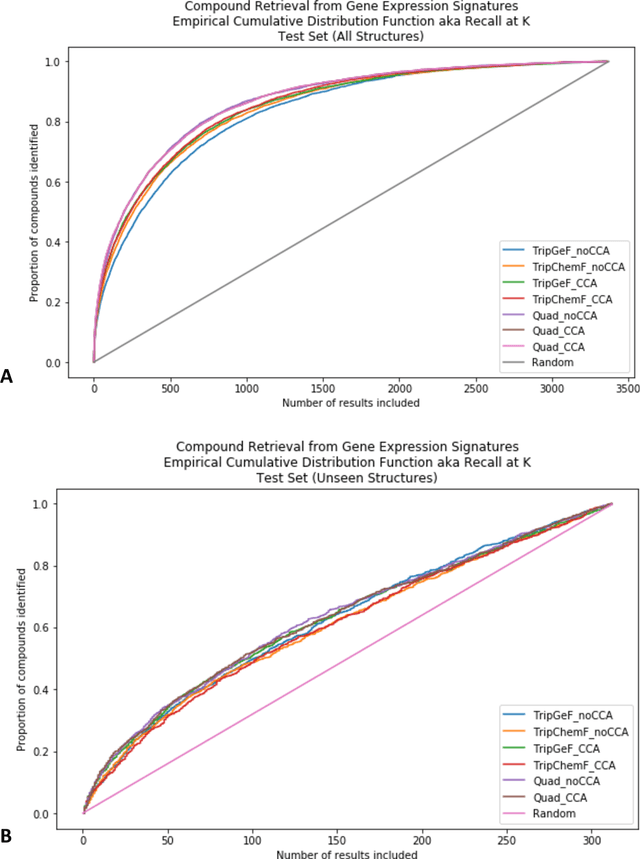
Abstract:Modeling the relationship between chemical structure and molecular activity is a key task in drug development and precision medicine. In this paper, we utilize a novel deep learning architecture to jointly train coordinated embeddings of chemical structures and transcriptional signatures. We do so by training neural networks in a coordinated manner such that learned chemical representations correlate most highly with the encodings of the transcriptional patterns they induce. We then test this approach by using held-out gene expression signatures as queries into embedding space to recover their corresponding compounds. We evaluate these embeddings' utility for small molecule prioritization on this new benchmark task. Our method outperforms a series of baselines, successfully generalizing to unseen transcriptional experiments, but still struggles to generalize to entirely unseen chemical structures.
Feature Robustness in Non-stationary Health Records: Caveats to Deployable Model Performance in Common Clinical Machine Learning Tasks
Aug 02, 2019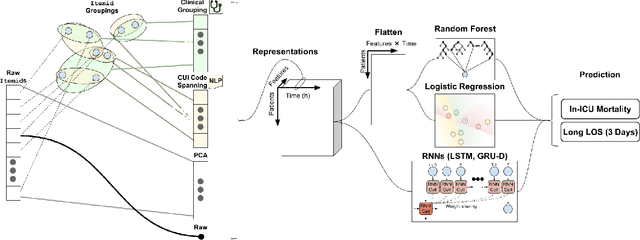
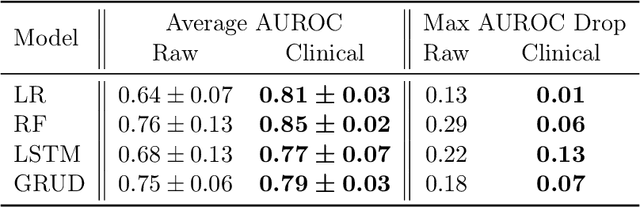
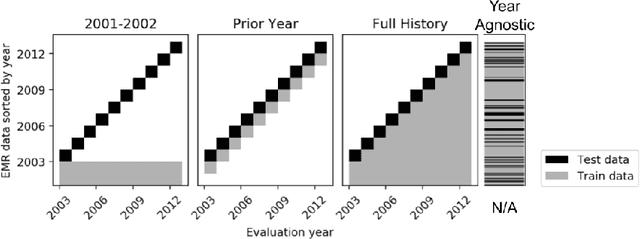
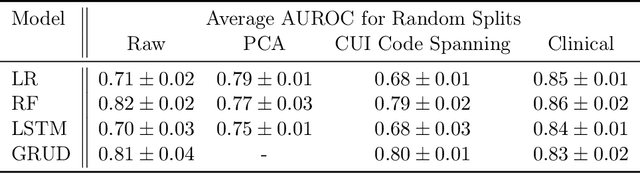
Abstract:When training clinical prediction models from electronic health records (EHRs), a key concern should be a model's ability to sustain performance over time when deployed, even as care practices, database systems, and population demographics evolve. Due to de-identification requirements, however, current experimental practices for public EHR benchmarks (such as the MIMIC-III critical care dataset) are time agnostic, assigning care records to train or test sets without regard for the actual dates of care. As a result, current benchmarks cannot assess how well models trained on one year generalise to another. In this work, we obtain a Limited Data Use Agreement to access year of care for each record in MIMIC and show that all tested state-of-the-art models decay in prediction quality when trained on historical data and tested on future data, particularly in response to a system-wide record-keeping change in 2008 (0.29 drop in AUROC for mortality prediction, 0.10 drop in AUROC for length-of-stay prediction with a random forest classifier). We further develop a simple yet effective mitigation strategy: by aggregating raw features into expert-defined clinical concepts, we see only a 0.06 drop in AUROC for mortality prediction and a 0.03 drop in AUROC for length-of-stay prediction. We demonstrate that this aggregation strategy outperforms other automatic feature preprocessing techniques aimed at increasing robustness to data drift. We release our aggregated representations and code to encourage more deployable clinical prediction models.
REflex: Flexible Framework for Relation Extraction in Multiple Domains
Jul 20, 2019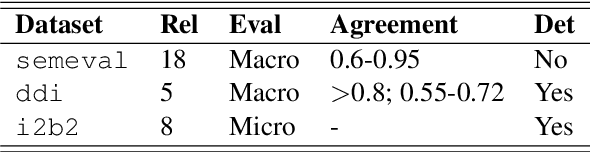
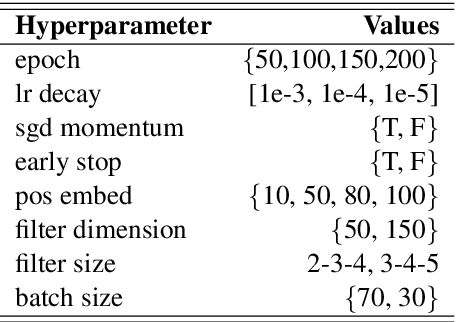
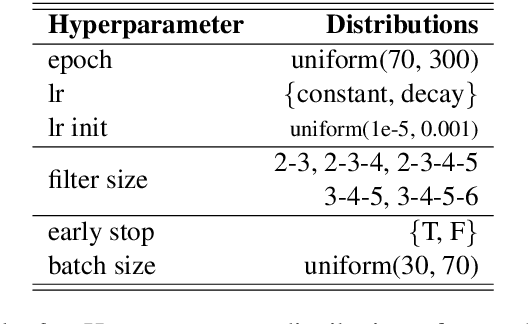
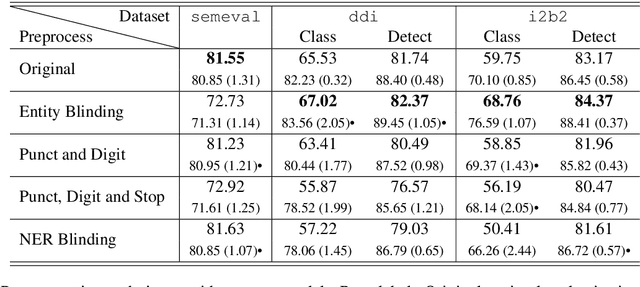
Abstract:Systematic comparison of methods for relation extraction (RE) is difficult because many experiments in the field are not described precisely enough to be completely reproducible and many papers fail to report ablation studies that would highlight the relative contributions of their various combined techniques. In this work, we build a unifying framework for RE, applying this on three highly used datasets (from the general, biomedical and clinical domains) with the ability to be extendable to new datasets. By performing a systematic exploration of modeling, pre-processing and training methodologies, we find that choices of pre-processing are a large contributor performance and that omission of such information can further hinder fair comparison. Other insights from our exploration allow us to provide recommendations for future research in this area.
MIMIC-Extract: A Data Extraction, Preprocessing, and Representation Pipeline for MIMIC-III
Jul 19, 2019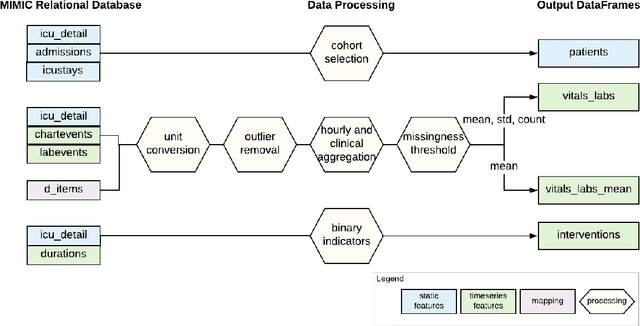
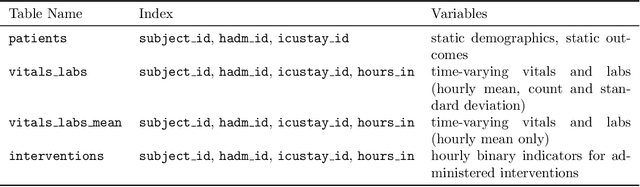
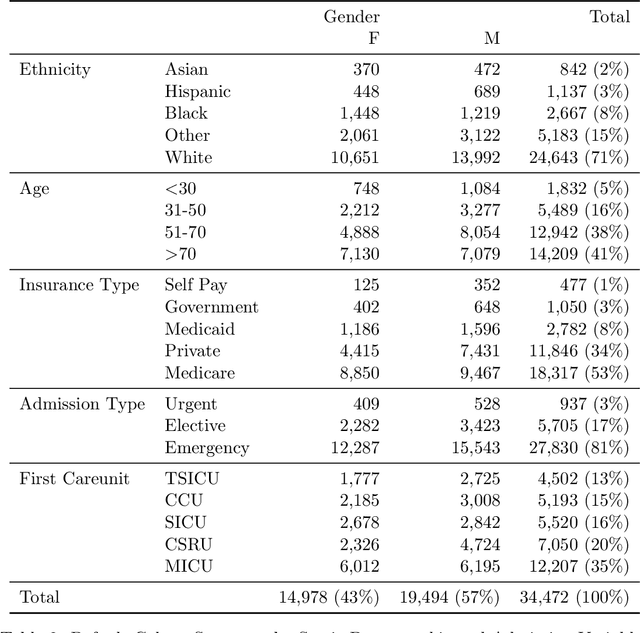
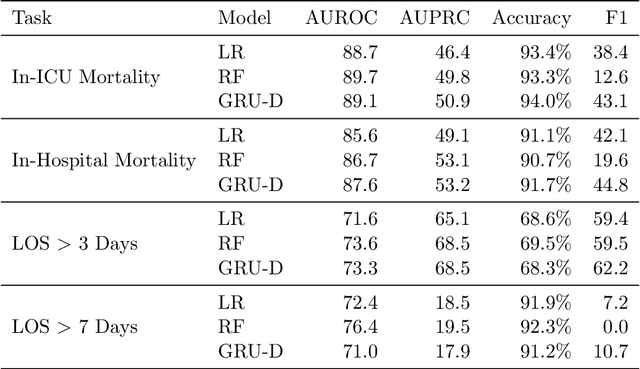
Abstract:Robust machine learning relies on access to data that can be used with standardized frameworks in important tasks and the ability to develop models whose performance can be reasonably reproduced. In machine learning for healthcare, the community faces reproducibility challenges due to a lack of publicly accessible data and a lack of standardized data processing frameworks. We present MIMIC-Extract, an open-source pipeline for transforming raw electronic health record (EHR) data for critical care patients contained in the publicly-available MIMIC-III database into dataframes that are directly usable in common machine learning pipelines. MIMIC-Extract addresses three primary challenges in making complex health records data accessible to the broader machine learning community. First, it provides standardized data processing functions, including unit conversion, outlier detection, and aggregating semantically equivalent features, thus accounting for duplication and reducing missingness. Second, it preserves the time series nature of clinical data and can be easily integrated into clinically actionable prediction tasks in machine learning for health. Finally, it is highly extensible so that other researchers with related questions can easily use the same pipeline. We demonstrate the utility of this pipeline by showcasing several benchmark tasks and baseline results.
Reproducibility in Machine Learning for Health
Jul 02, 2019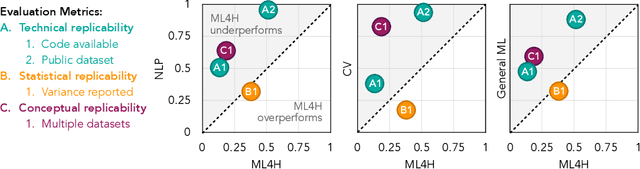
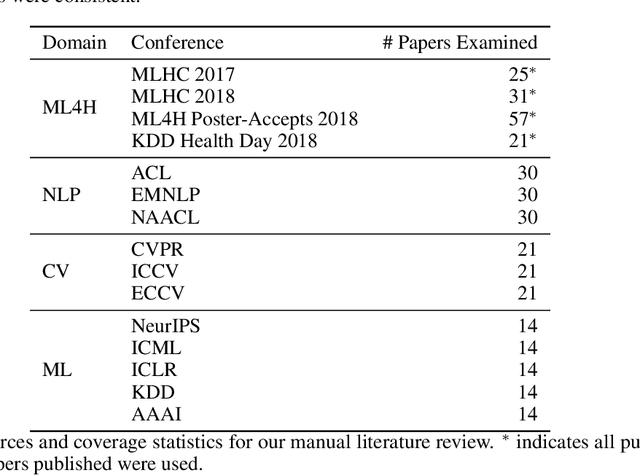
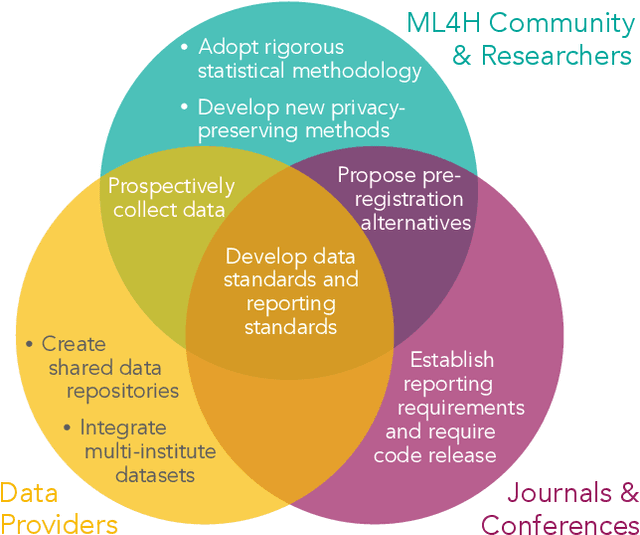
Abstract:Machine learning algorithms designed to characterize, monitor, and intervene on human health (ML4H) are expected to perform safely and reliably when operating at scale, potentially outside strict human supervision. This requirement warrants a stricter attention to issues of reproducibility than other fields of machine learning. In this work, we conduct a systematic evaluation of over 100 recently published ML4H research papers along several dimensions related to reproducibility. We find that the field of ML4H compares poorly to more established machine learning fields, particularly concerning data and code accessibility. Finally, drawing from success in other fields of science, we propose recommendations to data providers, academic publishers, and the ML4H research community in order to promote reproducible research moving forward.
Publicly Available Clinical BERT Embeddings
Apr 29, 2019
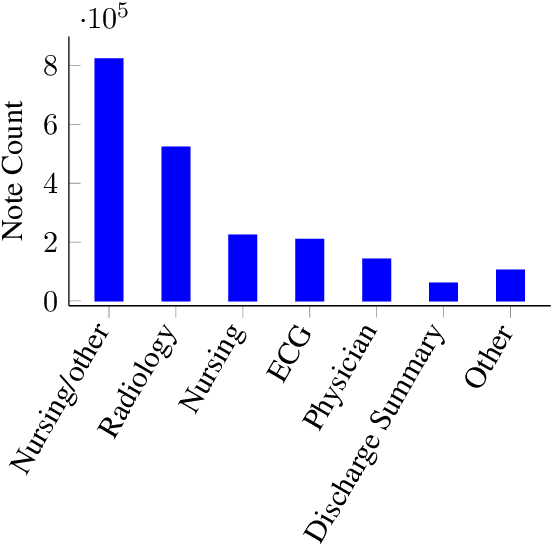
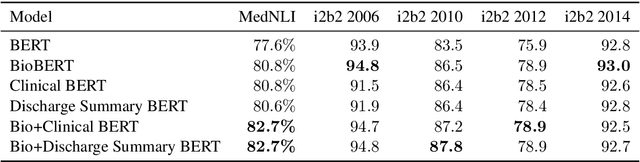
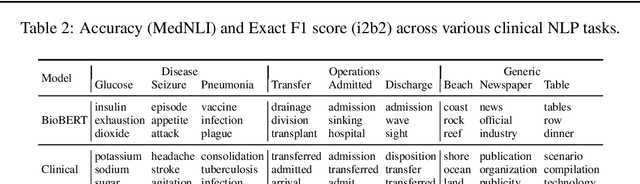
Abstract:Contextual word embedding models such as ELMo (Peters et al., 2018) and BERT (Devlin et al., 2018) have dramatically improved performance for many natural language processing (NLP) tasks in recent months. However, these models have been minimally explored on specialty corpora, such as clinical text; moreover, in the clinical domain, no publicly-available pre-trained BERT models yet exist. In this work, we address this need by exploring and releasing BERT models for clinical text: one for generic clinical text and another for discharge summaries specifically. We demonstrate that using a domain-specific model yields performance improvements on three common clinical NLP tasks as compared to nonspecific embeddings. These domain-specific models are not as performant on two clinical de-identification tasks, and argue that this is a natural consequence of the differences between de-identified source text and synthetically non de-identified task text.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge