Kwanhyung Lee
EMR-AGENT: Automating Cohort and Feature Extraction from EMR Databases
Oct 02, 2025Abstract:Machine learning models for clinical prediction rely on structured data extracted from Electronic Medical Records (EMRs), yet this process remains dominated by hardcoded, database-specific pipelines for cohort definition, feature selection, and code mapping. These manual efforts limit scalability, reproducibility, and cross-institutional generalization. To address this, we introduce EMR-AGENT (Automated Generalized Extraction and Navigation Tool), an agent-based framework that replaces manual rule writing with dynamic, language model-driven interaction to extract and standardize structured clinical data. Our framework automates cohort selection, feature extraction, and code mapping through interactive querying of databases. Our modular agents iteratively observe query results and reason over schema and documentation, using SQL not just for data retrieval but also as a tool for database observation and decision making. This eliminates the need for hand-crafted, schema-specific logic. To enable rigorous evaluation, we develop a benchmarking codebase for three EMR databases (MIMIC-III, eICU, SICdb), including both seen and unseen schema settings. Our results demonstrate strong performance and generalization across these databases, highlighting the feasibility of automating a process previously thought to require expert-driven design. The code will be released publicly at https://github.com/AITRICS/EMR-AGENT/tree/main. For a demonstration, please visit our anonymous demo page: https://anonymoususer-max600.github.io/EMR_AGENT/
Compact and De-biased Negative Instance Embedding for Multi-Instance Learning on Whole-Slide Image Classification
Feb 16, 2024



Abstract:Whole-slide image (WSI) classification is a challenging task because 1) patches from WSI lack annotation, and 2) WSI possesses unnecessary variability, e.g., stain protocol. Recently, Multiple-Instance Learning (MIL) has made significant progress, allowing for classification based on slide-level, rather than patch-level, annotations. However, existing MIL methods ignore that all patches from normal slides are normal. Using this free annotation, we introduce a semi-supervision signal to de-bias the inter-slide variability and to capture the common factors of variation within normal patches. Because our method is orthogonal to the MIL algorithm, we evaluate our method on top of the recently proposed MIL algorithms and also compare the performance with other semi-supervised approaches. We evaluate our method on two public WSI datasets including Camelyon-16 and TCGA lung cancer and demonstrate that our approach significantly improves the predictive performance of existing MIL algorithms and outperforms other semi-supervised algorithms. We release our code at https://github.com/AITRICS/pathology_mil.
Learning Missing Modal Electronic Health Records with Unified Multi-modal Data Embedding and Modality-Aware Attention
May 04, 2023Abstract:Electronic Health Record (EHR) provides abundant information through various modalities. However, learning multi-modal EHR is currently facing two major challenges, namely, 1) data embedding and 2) cases with missing modality. A lack of shared embedding function across modalities can discard the temporal relationship between different EHR modalities. On the other hand, most EHR studies are limited to relying only on EHR Times-series, and therefore, missing modality in EHR has not been well-explored. Therefore, in this study, we introduce a Unified Multi-modal Set Embedding (UMSE) and Modality-Aware Attention (MAA) with Skip Bottleneck (SB). UMSE treats all EHR modalities without a separate imputation module or error-prone carry-forward, whereas MAA with SB learns missing modal EHR with effective modality-aware attention. Our model outperforms other baseline models in mortality, vasopressor need, and intubation need prediction with the MIMIC-IV dataset.
Self-supervised predictive coding and multimodal fusion advance patient deterioration prediction in fine-grained time resolution
Oct 29, 2022Abstract:In the Emergency Department (ED), accurate prediction of critical events using Electronic Health Records (EHR) allows timely intervention and effective resource allocation. Though many studies have suggested automatic prediction methods, their coarse-grained time resolutions limit their practical usage. Therefore, in this study, we propose an hourly prediction method of critical events in ED, i.e., mortality and vasopressor need. Through extensive experiments, we show that both 1) bi-modal fusion between EHR text and time-series data and 2) self-supervised predictive regularization using L2 loss between normalized context vector and EHR future time-series data improve predictive performance, especially the far-future prediction. Our uni-modal/bi-modal/bi-modal self-supervision scored 0.846/0.877/0.897 (0.824/0.855/0.886) and 0.817/0.820/0.858 (0.807/0.81/0.855) with mortality (far-future mortality) and with vasopressor need (far-future vasopressor need) prediction data in AUROC, respectively.
Real-Time Seizure Detection using EEG: A Comprehensive Comparison of Recent Approaches under a Realistic Setting
Jan 21, 2022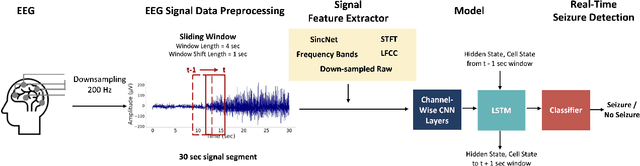
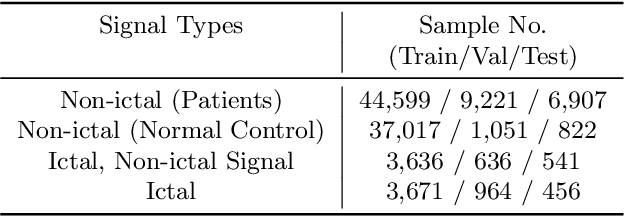
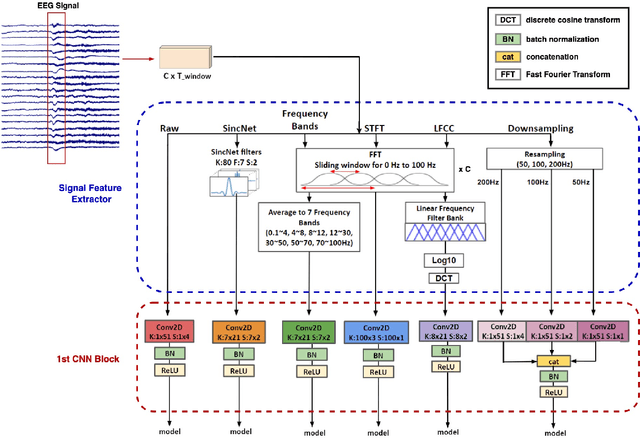
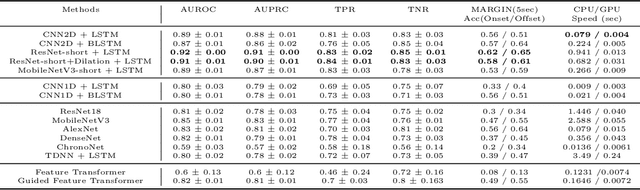
Abstract:Electroencephalogram (EEG) is an important diagnostic test that physicians use to record brain activity and detect seizures by monitoring the signals. There have been several attempts to detect seizures and abnormalities in EEG signals with modern deep learning models to reduce the clinical burden. However, they cannot be fairly compared against each other as they were tested in distinct experimental settings. Also, some of them are not trained in real-time seizure detection tasks, making it hard for on-device applications. Therefore in this work, for the first time, we extensively compare multiple state-of-the-art models and signal feature extractors in a real-time seizure detection framework suitable for real-world application, using various evaluation metrics including a new one we propose to evaluate more practical aspects of seizure detection models. Our code is available at https://github.com/AITRICS/EEG_real_time_seizure_detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge