Kirti Magudia
Diagnostic Impact of Cine Clips for Thyroid Nodule Assessment on Ultrasound
Feb 01, 2026Abstract:Background: Thyroid ultrasound is commonly performed using a combination of static images and cine clips (video recordings). However, the exact utility and impact of cine images remains unknown. This study aimed to evaluate the impact of cine imaging on accuracy and consistency of thyroid nodule assessment, using the American College of Radiology Thyroid Reporting and Data System (ACR TI-RADS). Methods: 50 benign and 50 malignant thyroid nodules with cytopathology results were included. A reader study with 4 specialty-trained radiologists was then conducted over 3 rounds, assessing only static images in the first two rounds and both static and cine images in the third round. TI-RADS scores and the consequent management recommendations were then evaluated by comparing them to the malignancy status of the nodules. Results: Mean sensitivity for malignancy detection was 0.65 for static images and 0.67 with both static and cine images (p>0.5). Specificity was 0.20 for static images and 0.22 with both static and cine images (p>0.5). Management recommendations were similar with and without cine images. Intrareader agreement on feature assignments remained consistent across all rounds, though TI-RADS point totals were slightly higher with cine images. Conclusion: The inclusion of cine imaging for thyroid nodule assessment on ultrasound did not significantly change diagnostic performance. Current practice guidelines, which do not mandate cine imaging, are sufficient for accurate diagnosis.
Language Models and Retrieval Augmented Generation for Automated Structured Data Extraction from Diagnostic Reports
Sep 18, 2024



Abstract:Purpose: To develop and evaluate an automated system for extracting structured clinical information from unstructured radiology and pathology reports using open-weights large language models (LMs) and retrieval augmented generation (RAG), and to assess the effects of model configuration variables on extraction performance. Methods and Materials: The study utilized two datasets: 7,294 radiology reports annotated for Brain Tumor Reporting and Data System (BT-RADS) scores and 2,154 pathology reports annotated for isocitrate dehydrogenase (IDH) mutation status. An automated pipeline was developed to benchmark the performance of various LMs and RAG configurations. The impact of model size, quantization, prompting strategies, output formatting, and inference parameters was systematically evaluated. Results: The best performing models achieved over 98% accuracy in extracting BT-RADS scores from radiology reports and over 90% for IDH mutation status extraction from pathology reports. The top model being medical fine-tuned llama3. Larger, newer, and domain fine-tuned models consistently outperformed older and smaller models. Model quantization had minimal impact on performance. Few-shot prompting significantly improved accuracy. RAG improved performance for complex pathology reports but not for shorter radiology reports. Conclusions: Open LMs demonstrate significant potential for automated extraction of structured clinical data from unstructured clinical reports with local privacy-preserving application. Careful model selection, prompt engineering, and semi-automated optimization using annotated data are critical for optimal performance. These approaches could be reliable enough for practical use in research workflows, highlighting the potential for human-machine collaboration in healthcare data extraction.
The RSNA Abdominal Traumatic Injury CT (RATIC) Dataset
May 30, 2024



Abstract:The RSNA Abdominal Traumatic Injury CT (RATIC) dataset is the largest publicly available collection of adult abdominal CT studies annotated for traumatic injuries. This dataset includes 4,274 studies from 23 institutions across 14 countries. The dataset is freely available for non-commercial use via Kaggle at https://www.kaggle.com/competitions/rsna-2023-abdominal-trauma-detection. Created for the RSNA 2023 Abdominal Trauma Detection competition, the dataset encourages the development of advanced machine learning models for detecting abdominal injuries on CT scans. The dataset encompasses detection and classification of traumatic injuries across multiple organs, including the liver, spleen, kidneys, bowel, and mesentery. Annotations were created by expert radiologists from the American Society of Emergency Radiology (ASER) and Society of Abdominal Radiology (SAR). The dataset is annotated at multiple levels, including the presence of injuries in three solid organs with injury grading, image-level annotations for active extravasations and bowel injury, and voxelwise segmentations of each of the potentially injured organs. With the release of this dataset, we hope to facilitate research and development in machine learning and abdominal trauma that can lead to improved patient care and outcomes.
Mixed Supervision of Histopathology Improves Prostate Cancer Classification from MRI
Dec 13, 2022



Abstract:Non-invasive prostate cancer detection from MRI has the potential to revolutionize patient care by providing early detection of clinically-significant disease (ISUP grade group >= 2), but has thus far shown limited positive predictive value. To address this, we present an MRI-based deep learning method for predicting clinically significant prostate cancer applicable to a patient population with subsequent ground truth biopsy results ranging from benign pathology to ISUP grade group~5. Specifically, we demonstrate that mixed supervision via diverse histopathological ground truth improves classification performance despite the cost of reduced concordance with image-based segmentation. That is, where prior approaches have utilized pathology results as ground truth derived from targeted biopsies and whole-mount prostatectomy to strongly supervise the localization of clinically significant cancer, our approach also utilizes weak supervision signals extracted from nontargeted systematic biopsies with regional localization to improve overall performance. Our key innovation is performing regression by distribution rather than simply by value, enabling use of additional pathology findings traditionally ignored by deep learning strategies. We evaluated our model on a dataset of 973 (testing n=160) multi-parametric prostate MRI exams collected at UCSF from 2015-2018 followed by MRI/ultrasound fusion (targeted) biopsy and systematic (nontargeted) biopsy of the prostate gland, demonstrating that deep networks trained with mixed supervision of histopathology can significantly exceed the performance of the Prostate Imaging-Reporting and Data System (PI-RADS) clinical standard for prostate MRI interpretation.
Federated Learning with Research Prototypes for Multi-Center MRI-based Detection of Prostate Cancer with Diverse Histopathology
Jun 11, 2022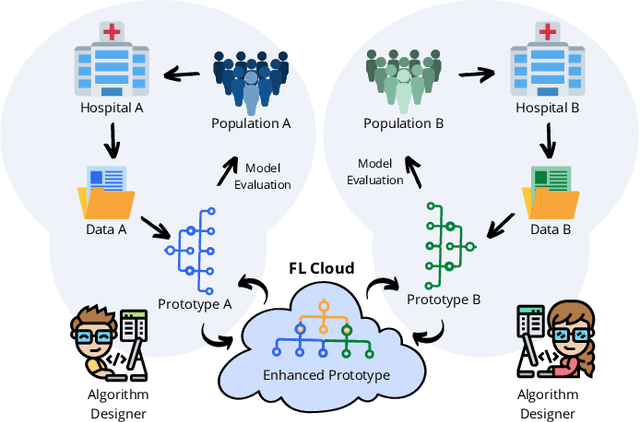
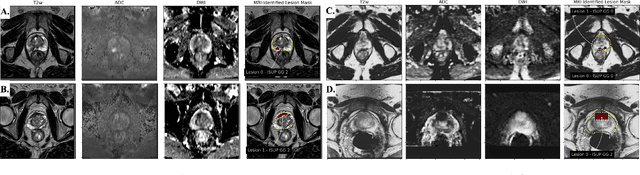
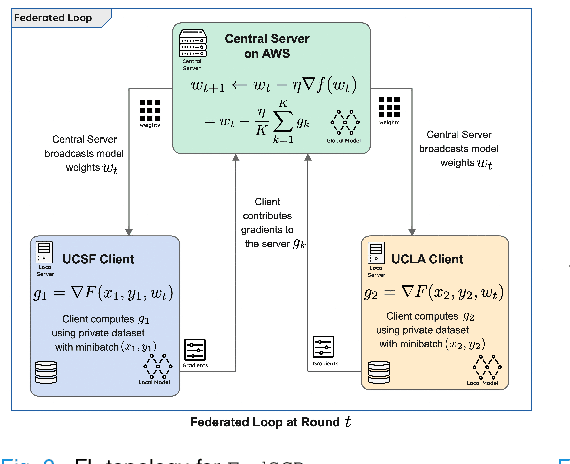
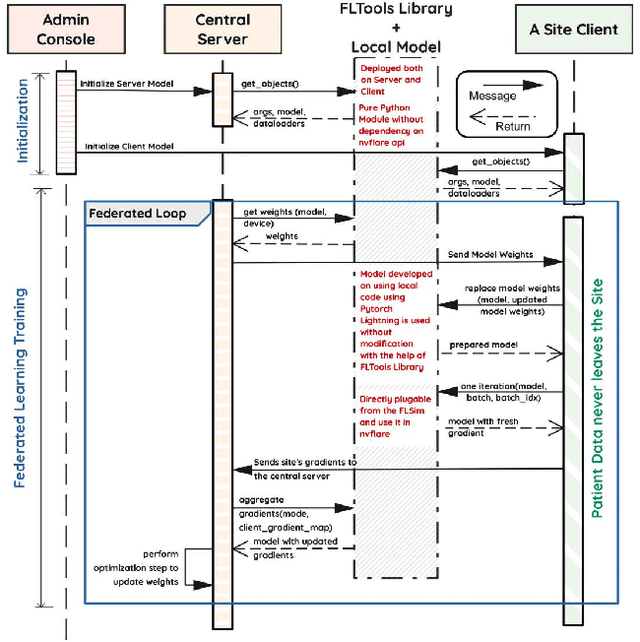
Abstract:Early prostate cancer detection and staging from MRI are extremely challenging tasks for both radiologists and deep learning algorithms, but the potential to learn from large and diverse datasets remains a promising avenue to increase their generalization capability both within- and across clinics. To enable this for prototype-stage algorithms, where the majority of existing research remains, in this paper we introduce a flexible federated learning framework for cross-site training, validation, and evaluation of deep prostate cancer detection algorithms. Our approach utilizes an abstracted representation of the model architecture and data, which allows unpolished prototype deep learning models to be trained without modification using the NVFlare federated learning framework. Our results show increases in prostate cancer detection and classification accuracy using a specialized neural network model and diverse prostate biopsy data collected at two University of California research hospitals, demonstrating the efficacy of our approach in adapting to different datasets and improving MR-biomarker discovery. We open-source our FLtools system, which can be easily adapted to other deep learning projects for medical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge