Kiho Im
An Intuitionistic Fuzzy Logic Driven UNet architecture: Application to Brain Image segmentation
Feb 04, 2026Abstract:Accurate segmentation of MRI brain images is essential for image analysis, diagnosis of neuro-logical disorders and medical image computing. In the deep learning approach, the convolutional neural networks (CNNs), especially UNet, are widely applied in medical image segmentation. However, it is difficult to deal with uncertainty due to the partial volume effect in brain images. To overcome this limitation, we propose an enhanced framework, named UNet with intuitionistic fuzzy logic (IF-UNet), which incorporates intuitionistic fuzzy logic into UNet. The model processes input data in terms of membership, nonmembership, and hesitation degrees, allowing it to better address tissue ambiguity resulting from partial volume effects and boundary uncertainties. The proposed architecture is evaluated on the Internet Brain Segmentation Repository (IBSR) dataset, and its performance is computed using accuracy, Dice coefficient, and intersection over union (IoU). Experimental results confirm that IF-UNet improves segmentation quality with handling uncertainty in brain images.
Fast Multi-Stack Slice-to-Volume Reconstruction via Multi-Scale Unrolled Optimization
Jan 12, 2026Abstract:Fully convolutional networks have become the backbone of modern medical imaging due to their ability to learn multi-scale representations and perform end-to-end inference. Yet their potential for slice-to-volume reconstruction (SVR), the task of jointly estimating 3D anatomy and slice poses from misaligned 2D acquisitions, remains underexplored. We introduce a fast convolutional framework that fuses multiple orthogonal 2D slice stacks to recover coherent 3D structure and refines slice alignment through lightweight model-based optimization. Applied to fetal brain MRI, our approach reconstructs high-quality 3D volumes in under 10s, with 1s slice registration and accuracy on par with state-of-the-art iterative SVR pipelines, offering more than speedup. The framework uses non-rigid displacement fields to represent transformations, generalizing to other SVR problems like fetal body and placental MRI. Additionally, the fast inference time paves the way for real-time, scanner-side volumetric feedback during MRI acquisition.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


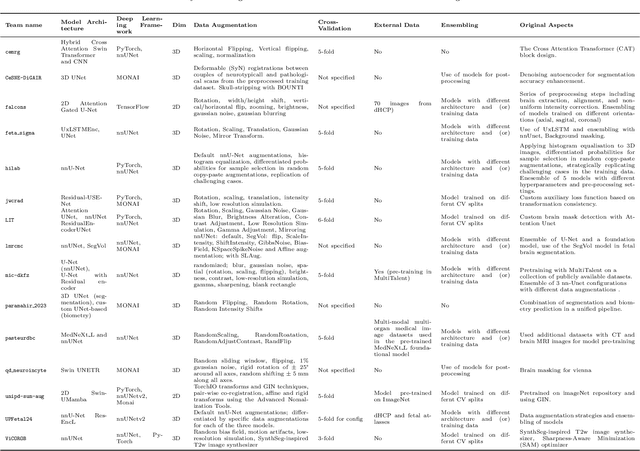
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge