Kevin S. Xu
LIT-LVM: Structured Regularization for Interaction Terms in Linear Predictors using Latent Variable Models
Jun 18, 2025Abstract:Some of the simplest, yet most frequently used predictors in statistics and machine learning use weighted linear combinations of features. Such linear predictors can model non-linear relationships between features by adding interaction terms corresponding to the products of all pairs of features. We consider the problem of accurately estimating coefficients for interaction terms in linear predictors. We hypothesize that the coefficients for different interaction terms have an approximate low-dimensional structure and represent each feature by a latent vector in a low-dimensional space. This low-dimensional representation can be viewed as a structured regularization approach that further mitigates overfitting in high-dimensional settings beyond standard regularizers such as the lasso and elastic net. We demonstrate that our approach, called LIT-LVM, achieves superior prediction accuracy compared to elastic net and factorization machines on a wide variety of simulated and real data, particularly when the number of interaction terms is high compared to the number of samples. LIT-LVM also provides low-dimensional latent representations for features that are useful for visualizing and analyzing their relationships.
Spectral clustering for dependent community Hawkes process models of temporal networks
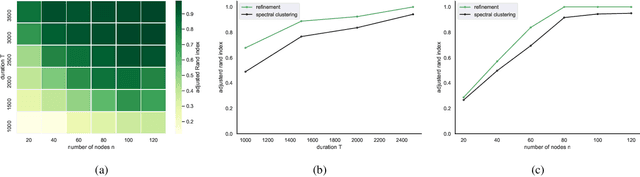
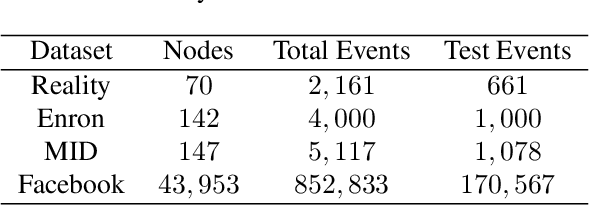
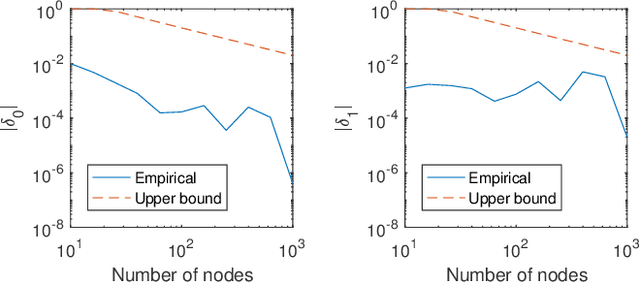
May 28, 2025Abstract:Temporal networks observed continuously over time through timestamped relational events data are commonly encountered in application settings including online social media communications, financial transactions, and international relations. Temporal networks often exhibit community structure and strong dependence patterns among node pairs. This dependence can be modeled through mutual excitations, where an interaction event from a sender to a receiver node increases the possibility of future events among other node pairs. We provide statistical results for a class of models that we call dependent community Hawkes (DCH) models, which combine the stochastic block model with mutually exciting Hawkes processes for modeling both community structure and dependence among node pairs, respectively. We derive a non-asymptotic upper bound on the misclustering error of spectral clustering on the event count matrix as a function of the number of nodes and communities, time duration, and the amount of dependence in the model. Our result leverages recent results on bounding an appropriate distance between a multivariate Hawkes process count vector and a Gaussian vector, along with results from random matrix theory. We also propose a DCH model that incorporates only self and reciprocal excitation along with highly scalable parameter estimation using a Generalized Method of Moments (GMM) estimator that we demonstrate to be consistent for growing network size and time duration.
A Latent Space Model for HLA Compatibility Networks in Kidney Transplantation
Nov 04, 2022



Abstract:Kidney transplantation is the preferred treatment for people suffering from end-stage renal disease. Successful kidney transplants still fail over time, known as graft failure; however, the time to graft failure, or graft survival time, can vary significantly between different recipients. A significant biological factor affecting graft survival times is the compatibility between the human leukocyte antigens (HLAs) of the donor and recipient. We propose to model HLA compatibility using a network, where the nodes denote different HLAs of the donor and recipient, and edge weights denote compatibilities of the HLAs, which can be positive or negative. The network is indirectly observed, as the edge weights are estimated from transplant outcomes rather than directly observed. We propose a latent space model for such indirectly-observed weighted and signed networks. We demonstrate that our latent space model can not only result in more accurate estimates of HLA compatibilities, but can also be incorporated into survival analysis models to improve accuracy for the downstream task of predicting graft survival times.
A Mutually Exciting Latent Space Hawkes Process Model for Continuous-time Networks
May 19, 2022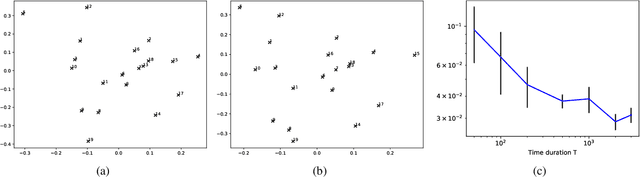
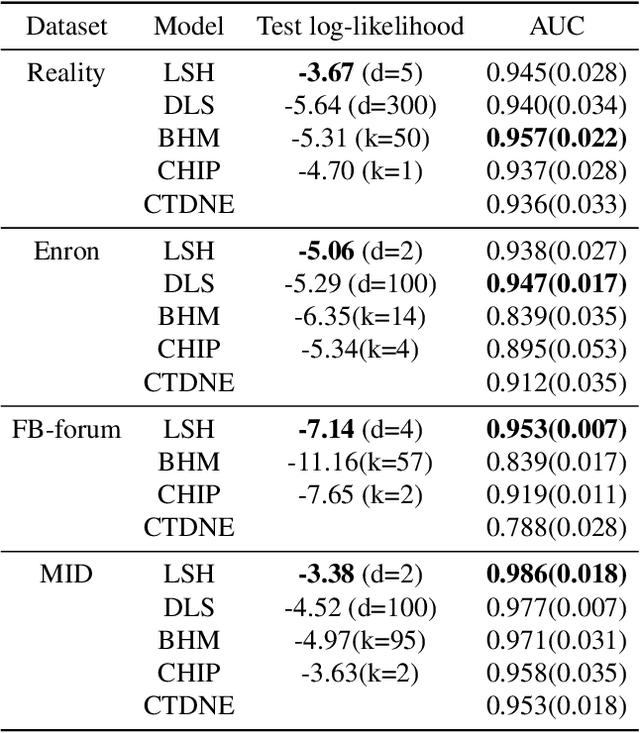
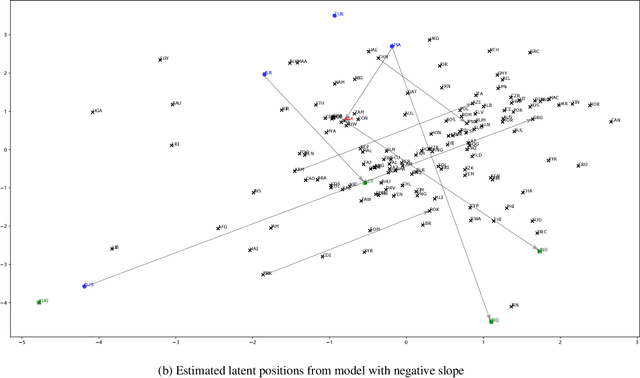
Abstract:Networks and temporal point processes serve as fundamental building blocks for modeling complex dynamic relational data in various domains. We propose the latent space Hawkes (LSH) model, a novel generative model for continuous-time networks of relational events, using a latent space representation for nodes. We model relational events between nodes using mutually exciting Hawkes processes with baseline intensities dependent upon the distances between the nodes in the latent space and sender and receiver specific effects. We propose an alternating minimization algorithm to jointly estimate the latent positions of the nodes and other model parameters. We demonstrate that our proposed LSH model can replicate many features observed in real temporal networks including reciprocity and transitivity, while also achieves superior prediction accuracy and provides more interpretability compared to existing models.
The Multivariate Community Hawkes Model for Dependent Relational Events in Continuous-time Networks
May 02, 2022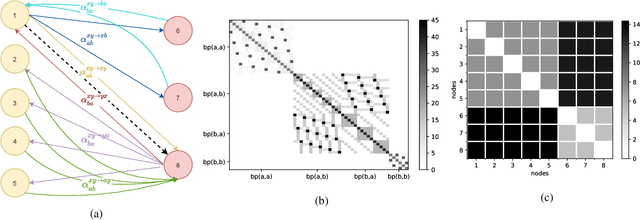
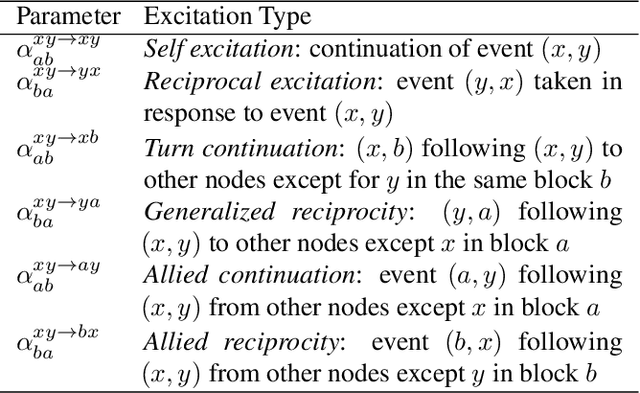
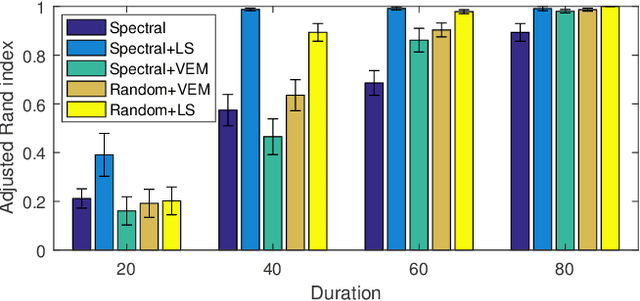
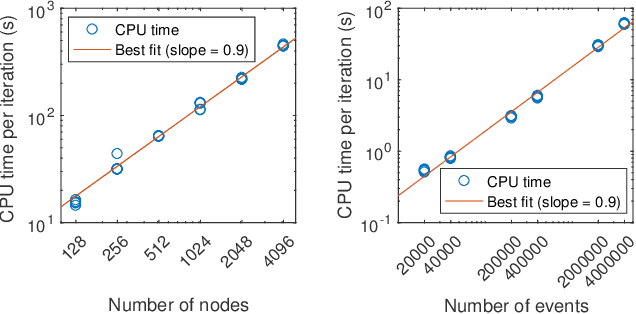
Abstract:The stochastic block model (SBM) is one of the most widely used generative models for network data. Many continuous-time dynamic network models are built upon the same assumption as the SBM: edges or events between all pairs of nodes are conditionally independent given the block or community memberships, which prevents them from reproducing higher-order motifs such as triangles that are commonly observed in real networks. We propose the multivariate community Hawkes (MULCH) model, an extremely flexible community-based model for continuous-time networks that introduces dependence between node pairs using structured multivariate Hawkes processes. We fit the model using a spectral clustering and likelihood-based local refinement procedure. We find that our proposed MULCH model is far more accurate than existing models both for predictive and generative tasks.
Learning User Embeddings from Temporal Social Media Data: A Survey
May 17, 2021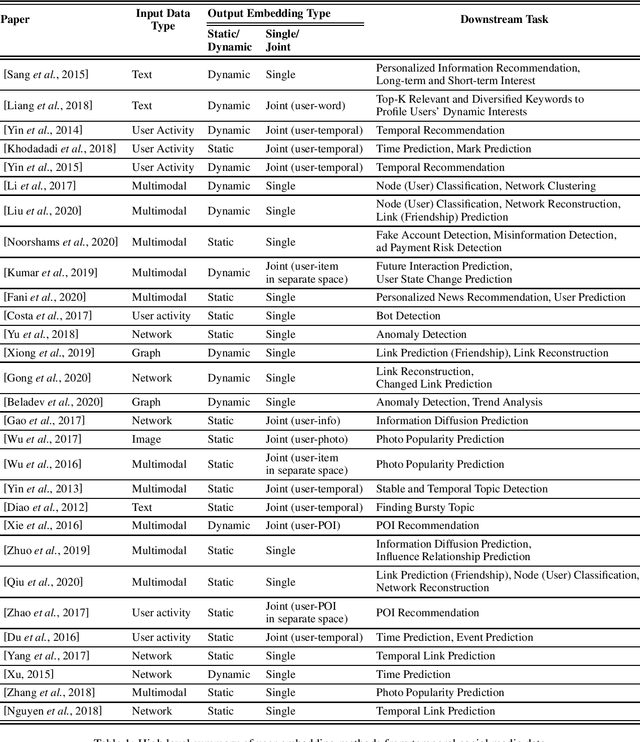
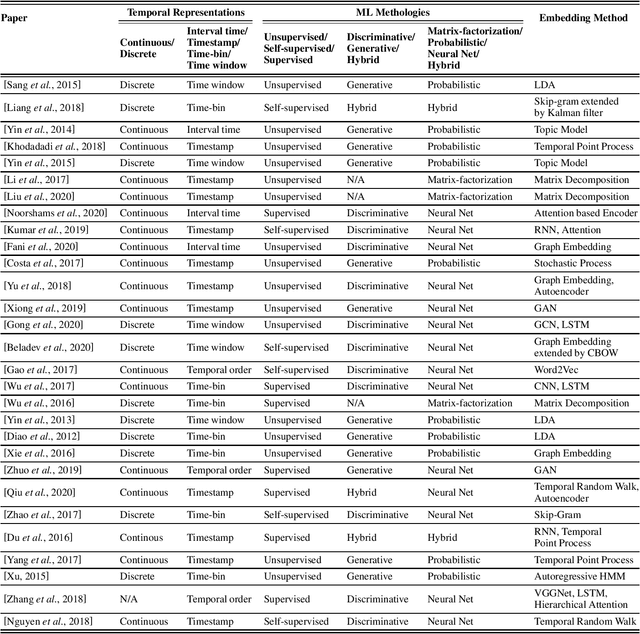
Abstract:User-generated data on social media contain rich information about who we are, what we like and how we make decisions. In this paper, we survey representative work on learning a concise latent user representation (a.k.a. user embedding) that can capture the main characteristics of a social media user. The learned user embeddings can later be used to support different downstream user analysis tasks such as personality modeling, suicidal risk assessment and purchase decision prediction. The temporal nature of user-generated data on social media has largely been overlooked in much of the existing user embedding literature. In this survey, we focus on research that bridges the gap by incorporating temporal/sequential information in user representation learning. We categorize relevant papers along several key dimensions, identify limitations in the current work and suggest future research directions.
Predicting Kidney Transplant Survival using Multiple Feature Representations for HLAs
Mar 04, 2021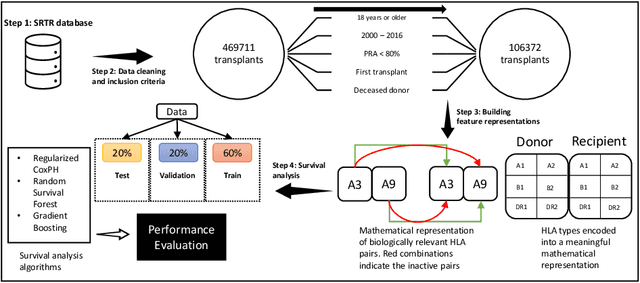
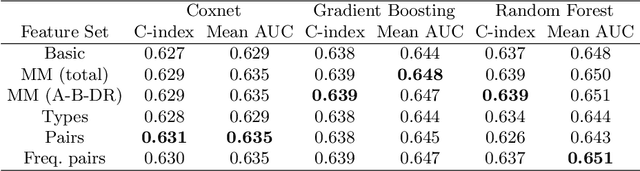
Abstract:Kidney transplantation can significantly enhance living standards for people suffering from end-stage renal disease. A significant factor that affects graft survival time (the time until the transplant fails and the patient requires another transplant) for kidney transplantation is the compatibility of the Human Leukocyte Antigens (HLAs) between the donor and recipient. In this paper, we propose new biologically-relevant feature representations for incorporating HLA information into machine learning-based survival analysis algorithms. We evaluate our proposed HLA feature representations on a database of over 100,000 transplants and find that they improve prediction accuracy by about 1%, modest at the patient level but potentially significant at a societal level. Accurate prediction of survival times can improve transplant survival outcomes, enabling better allocation of donors to recipients and reducing the number of re-transplants due to graft failure with poorly matched donors.
Consistent Community Detection in Continuous-Time Networks of Relational Events
Aug 19, 2019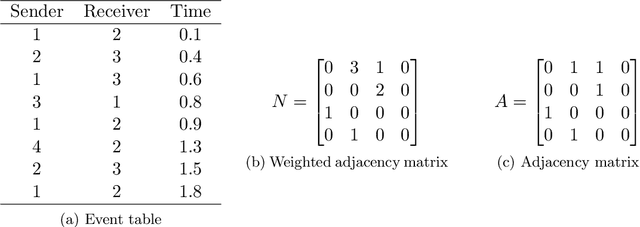
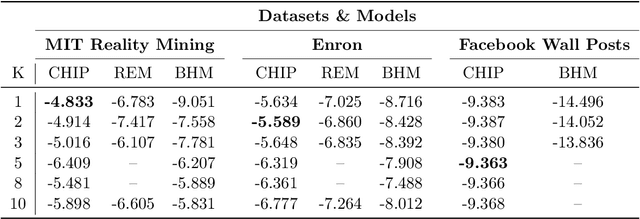
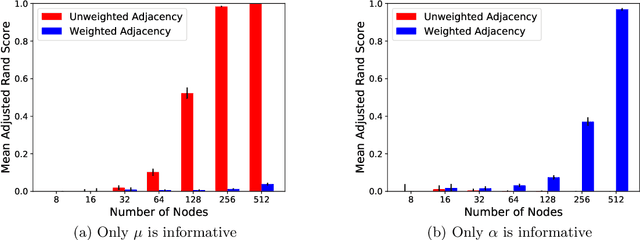
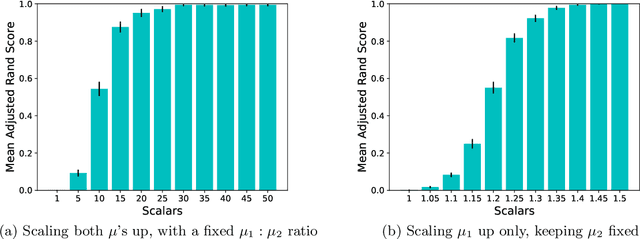
Abstract:In many application settings involving networks, such as messages between users of an on-line social network or transactions between traders in financial markets, the observed data are in the form of relational events with timestamps, which form a continuous-time network. We propose the Community Hawkes Independent Pairs (CHIP) model for community detection on such timestamped relational event data. We demonstrate that applying spectral clustering to adjacency matrices constructed from relational events generated by the CHIP model provides consistent community detection for a growing number of nodes. In particular, we obtain explicit non-asymptotic upper bounds on the misclustering rates based on the separation conditions required on the parameters of the model for consistent community detection. We also develop consistent and computationally efficient estimators for the parameters of the model. We demonstrate that our proposed CHIP model and estimation procedure scales to large networks with tens of thousands of nodes and provides superior fits compared to existing continuous-time network models on several real networks.
Block CUR: Decomposing Matrices using Groups of Columns
Jul 09, 2018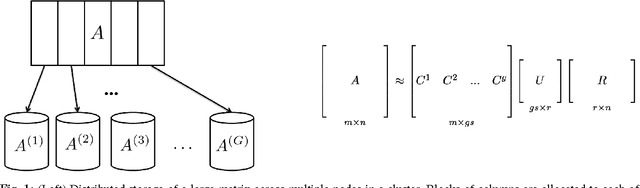
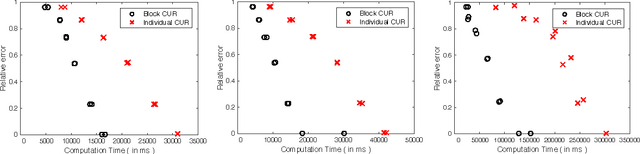
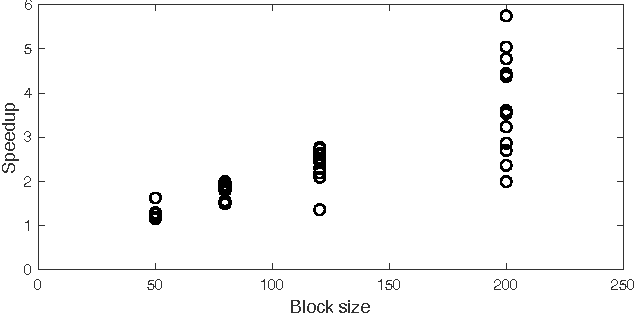
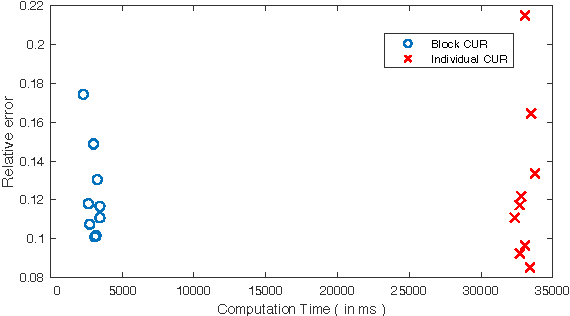
Abstract:A common problem in large-scale data analysis is to approximate a matrix using a combination of specifically sampled rows and columns, known as CUR decomposition. Unfortunately, in many real-world environments, the ability to sample specific individual rows or columns of the matrix is limited by either system constraints or cost. In this paper, we consider matrix approximation by sampling predefined \emph{blocks} of columns (or rows) from the matrix. We present an algorithm for sampling useful column blocks and provide novel guarantees for the quality of the approximation. This algorithm has application in problems as diverse as biometric data analysis to distributed computing. We demonstrate the effectiveness of the proposed algorithms for computing the Block CUR decomposition of large matrices in a distributed setting with multiple nodes in a compute cluster, where such blocks correspond to columns (or rows) of the matrix stored on the same node, which can be retrieved with much less overhead than retrieving individual columns stored across different nodes. In the biometric setting, the rows correspond to different users and columns correspond to users' biometric reaction to external stimuli, {\em e.g.,}~watching video content, at a particular time instant. There is significant cost in acquiring each user's reaction to lengthy content so we sample a few important scenes to approximate the biometric response. An individual time sample in this use case cannot be queried in isolation due to the lack of context that caused that biometric reaction. Instead, collections of time segments ({\em i.e.,} blocks) must be presented to the user. The practical application of these algorithms is shown via experimental results using real-world user biometric data from a content testing environment.
The Block Point Process Model for Continuous-Time Event-Based Dynamic Networks
Nov 29, 2017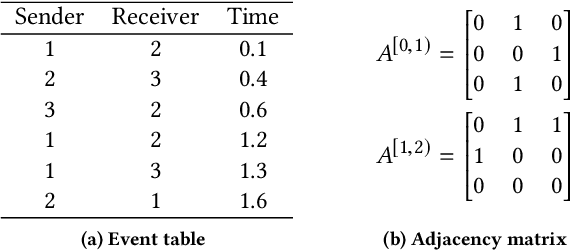
Abstract:Many application settings involve the analysis of timestamped relations or events between a set of entities, e.g. messages between users of an on-line social network. Static and discrete-time network models are typically used as analysis tools in these settings; however, they discard a significant amount of information by aggregating events over time to form network snapshots. In this paper, we introduce a block point process model (BPPM) for dynamic networks evolving in continuous time in the form of events at irregular time intervals. The BPPM is inspired by the well-known stochastic block model (SBM) for static networks and is a simpler version of the recently-proposed Hawkes infinite relational model (IRM). We show that networks generated by the BPPM follow an SBM in the limit of a growing number of nodes and leverage this property to develop an efficient inference procedure for the BPPM. We fit the BPPM to several real network data sets, including a Facebook network with over 3, 500 nodes and 130, 000 events, several orders of magnitude larger than the Hawkes IRM and other existing point process network models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge