Junhua Gu
MPCM-Net: Multi-scale network integrates partial attention convolution with Mamba for ground-based cloud image segmentation
Nov 12, 2025Abstract:Ground-based cloud image segmentation is a critical research domain for photovoltaic power forecasting. Current deep learning approaches primarily focus on encoder-decoder architectural refinements. However, existing methodologies exhibit several limitations:(1)they rely on dilated convolutions for multi-scale context extraction, lacking the partial feature effectiveness and interoperability of inter-channel;(2)attention-based feature enhancement implementations neglect accuracy-throughput balance; and (3)the decoder modifications fail to establish global interdependencies among hierarchical local features, limiting inference efficiency. To address these challenges, we propose MPCM-Net, a Multi-scale network that integrates Partial attention Convolutions with Mamba architectures to enhance segmentation accuracy and computational efficiency. Specifically, the encoder incorporates MPAC, which comprises:(1)a MPC block with ParCM and ParSM that enables global spatial interaction across multi-scale cloud formations, and (2)a MPA block combining ParAM and ParSM to extract discriminative features with reduced computational complexity. On the decoder side, a M2B is employed to mitigate contextual loss through a SSHD that maintains linear complexity while enabling deep feature aggregation across spatial and scale dimensions. As a key contribution to the community, we also introduce and release a dataset CSRC, which is a clear-label, fine-grained segmentation benchmark designed to overcome the critical limitations of existing public datasets. Extensive experiments on CSRC demonstrate the superior performance of MPCM-Net over state-of-the-art methods, achieving an optimal balance between segmentation accuracy and inference speed. The dataset and source code will be available at https://github.com/she1110/CSRC.
Filling the Missings: Spatiotemporal Data Imputation by Conditional Diffusion
Jun 08, 2025Abstract:Missing data in spatiotemporal systems presents a significant challenge for modern applications, ranging from environmental monitoring to urban traffic management. The integrity of spatiotemporal data often deteriorates due to hardware malfunctions and software failures in real-world deployments. Current approaches based on machine learning and deep learning struggle to model the intricate interdependencies between spatial and temporal dimensions effectively and, more importantly, suffer from cumulative errors during the data imputation process, which propagate and amplify through iterations. To address these limitations, we propose CoFILL, a novel Conditional Diffusion Model for spatiotemporal data imputation. CoFILL builds on the inherent advantages of diffusion models to generate high-quality imputations without relying on potentially error-prone prior estimates. It incorporates an innovative dual-stream architecture that processes temporal and frequency domain features in parallel. By fusing these complementary features, CoFILL captures both rapid fluctuations and underlying patterns in the data, which enables more robust imputation. The extensive experiments reveal that CoFILL's noise prediction network successfully transforms random noise into meaningful values that align with the true data distribution. The results also show that CoFILL outperforms state-of-the-art methods in imputation accuracy. The source code is publicly available at https://github.com/joyHJL/CoFILL.
A systematic evaluation of methods for cell phenotype classification using single-cell RNA sequencing data
Oct 01, 2021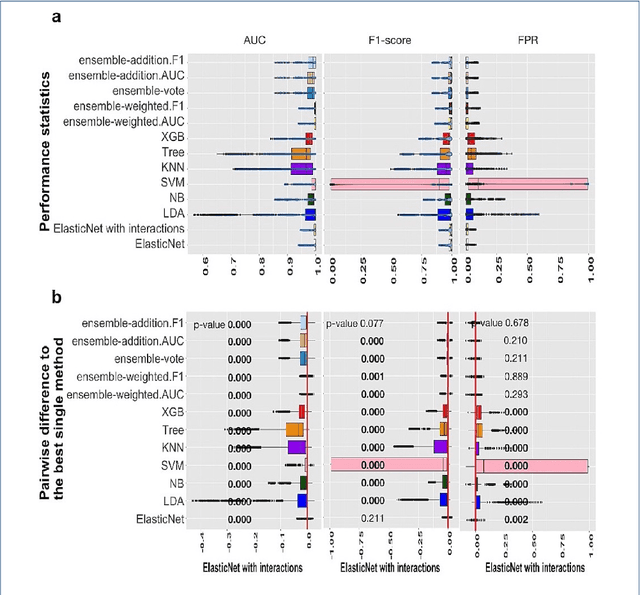
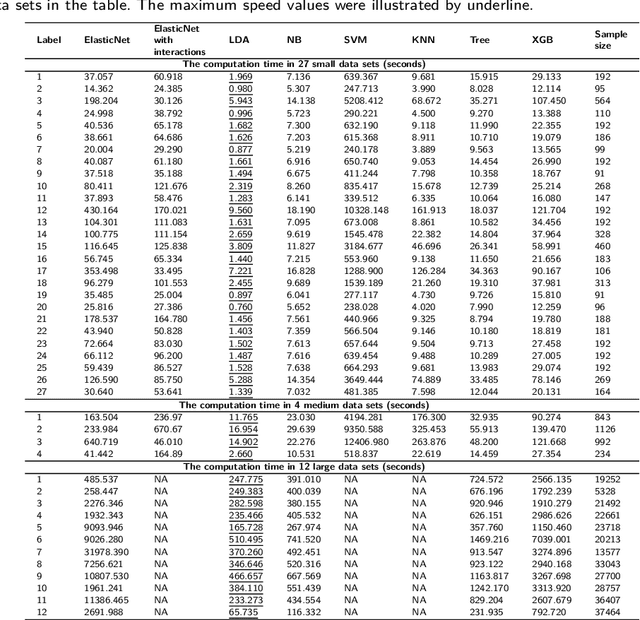
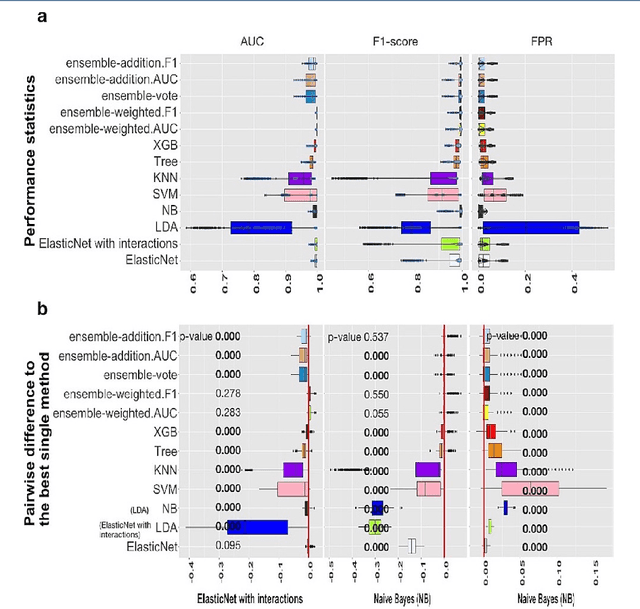
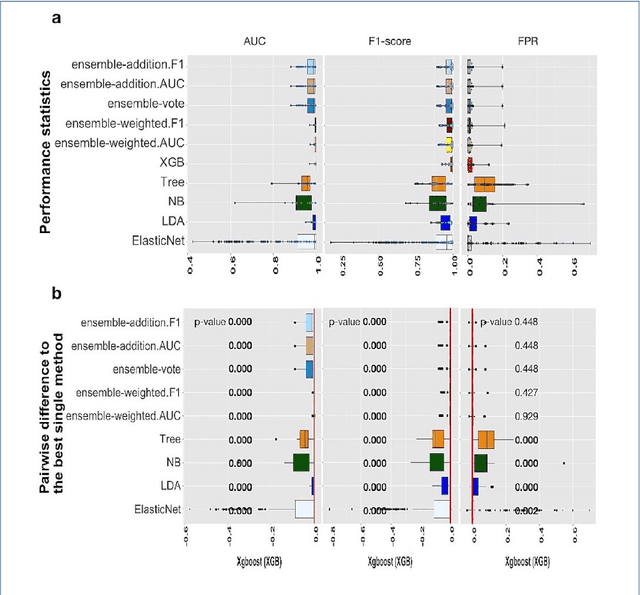
Abstract:Background: Single-cell RNA sequencing (scRNA-seq) yields valuable insights about gene expression and gives critical information about complex tissue cellular composition. In the analysis of single-cell RNA sequencing, the annotations of cell subtypes are often done manually, which is time-consuming and irreproducible. Garnett is a cell-type annotation software based the on elastic net method. Besides cell-type annotation, supervised machine learning methods can also be applied to predict other cell phenotypes from genomic data. Despite the popularity of such applications, there is no existing study to systematically investigate the performance of those supervised algorithms in various sizes of scRNA-seq data sets. Methods and Results: This study evaluates 13 popular supervised machine learning algorithms to classify cell phenotypes, using published real and simulated data sets with diverse cell sizes. The benchmark contained two parts. In the first part, we used real data sets to assess the popular supervised algorithms' computing speed and cell phenotype classification performance. The classification performances were evaluated using AUC statistics, F1-score, precision, recall, and false-positive rate. In the second part, we evaluated gene selection performance using published simulated data sets with a known list of real genes. Conclusion: The study outcomes showed that ElasticNet with interactions performed best in small and medium data sets. NB was another appropriate method for medium data sets. In large data sets, XGB works excellent. Ensemble algorithms were not significantly superior to individual machine learning methods. Adding interactions to ElasticNet can help, and the improvement was significant in small data sets.
Separating the EoR Signal with a Convolutional Denoising Autoencoder: A Deep-learning-based Method
Mar 14, 2019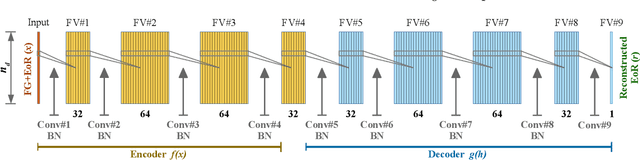
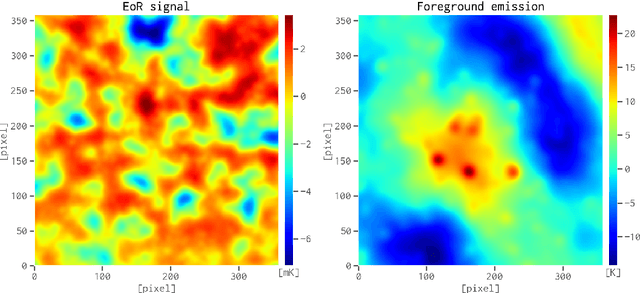
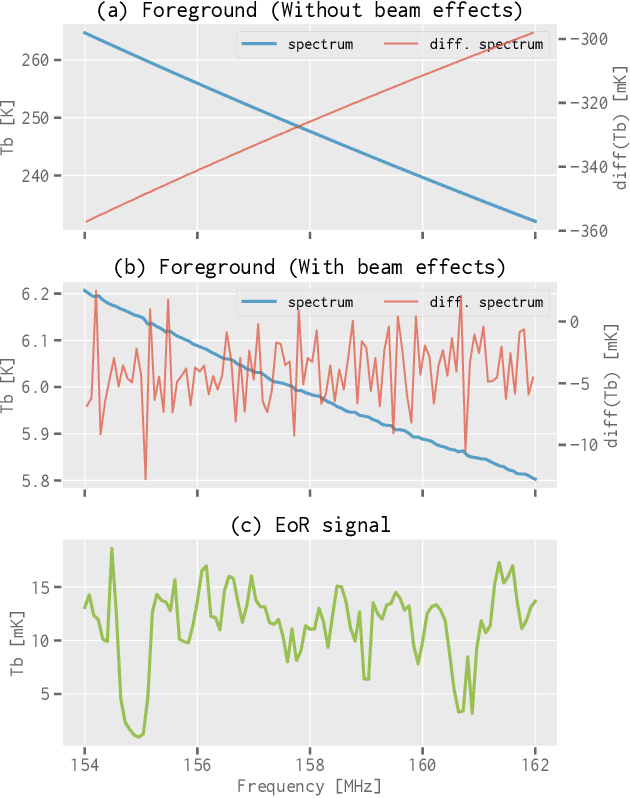
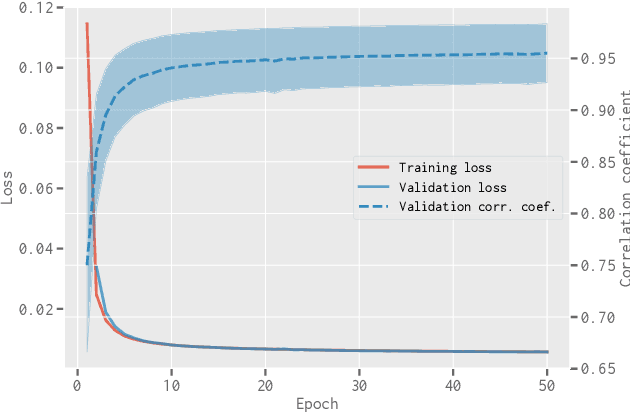
Abstract:When applying the foreground removal methods to uncover the faint cosmological signal from the epoch of reionization (EoR), the foreground spectra are assumed to be smooth. However, this assumption can be seriously violated in practice since the unresolved or mis-subtracted foreground sources, which are further complicated by the frequency-dependent beam effects of interferometers, will generate significant fluctuations along the frequency dimension. To address this issue, we propose a novel deep-learning-based method that uses a 9-layer convolutional denoising autoencoder (CDAE) to separate the EoR signal. After being trained on the SKA images simulated with realistic beam effects, the CDAE achieves excellent performance as the mean correlation coefficient ($\bar{\rho}$) between the reconstructed and input EoR signals reaches $0.929 \pm 0.045$. In comparison, the two representative traditional methods, namely the polynomial fitting method and the continuous wavelet transform method, both have difficulties in modelling and removing the foreground emission complicated with the beam effects, yielding only $\bar{\rho}_{\text{poly}} = 0.296 \pm 0.121$ and $\bar{\rho}_{\text{cwt}} = 0.198 \pm 0.160$, respectively. We conclude that, by hierarchically learning sophisticated features through multiple convolutional layers, the CDAE is a powerful tool that can be used to overcome the complicated beam effects and accurately separate the EoR signal. Our results also exhibit the great potential of deep-learning-based methods in future EoR experiments.
* 10 pages, 9 figures; minor text updates to match the MNRAS published version
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge