Julien Cohen-Adad
Unsupervised domain adaptation for medical imaging segmentation with self-ensembling
Nov 14, 2018
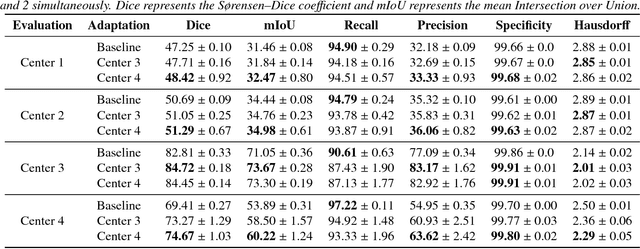
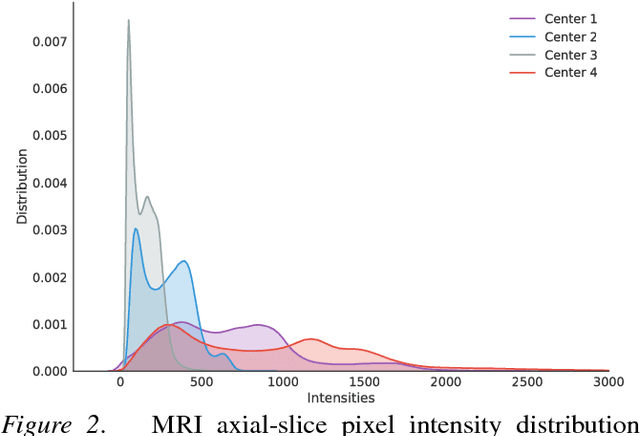
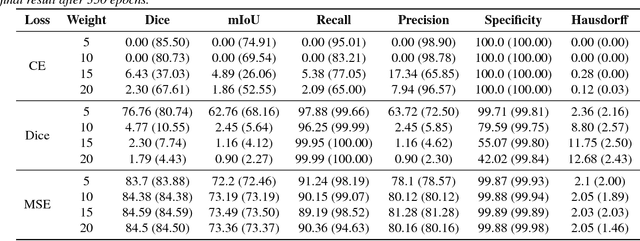
Abstract:Recent deep learning methods for the medical imaging domain have reached state-of-the-art results and even surpassed human judgment in several tasks. Those models, however, when trained to reduce the empirical risk on a single domain, fail to generalize when applied on other domains, a very common scenario on medical imaging due to the variability of images and anatomical structures, even across the same imaging modality. In this work, we extend the method of unsupervised domain adaptation using self-ensembling for the semantic segmentation task and explore multiple facets of the method on a realistic small data regime using a publicly available magnetic resonance (MRI) dataset. Through an extensive evaluation, we show that self-ensembling can indeed improve the generalization of the models even when using a small amount of unlabelled data.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018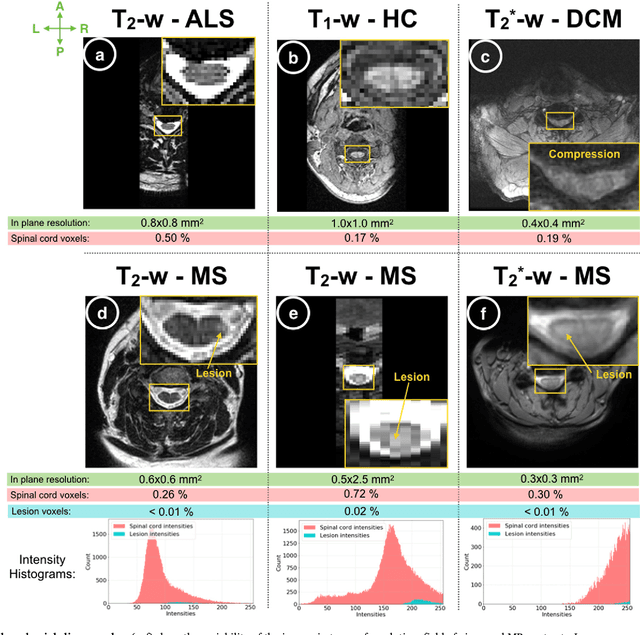
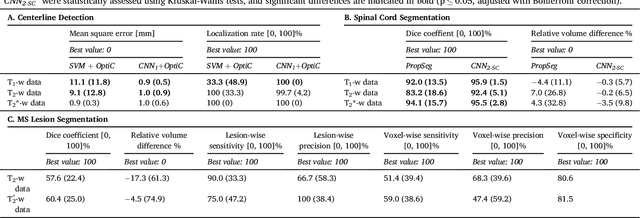
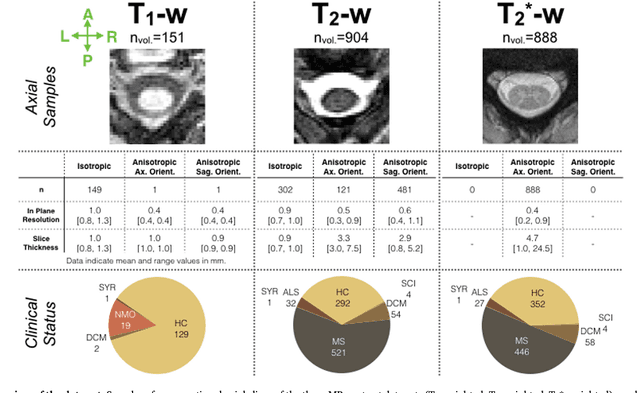
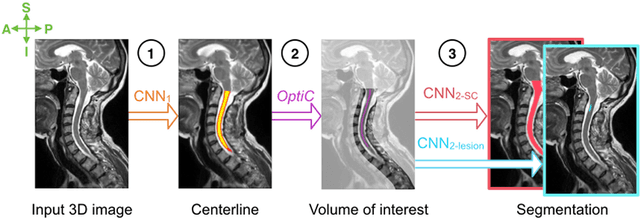
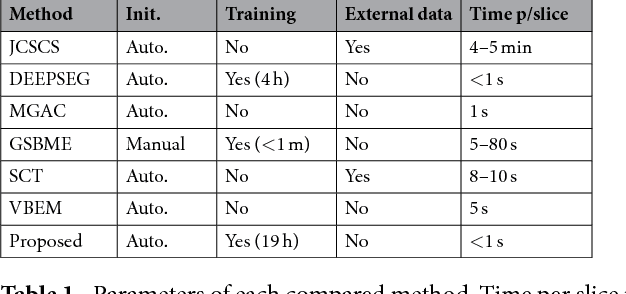
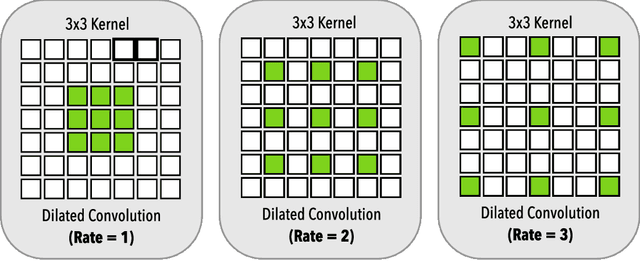
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
Deep semi-supervised segmentation with weight-averaged consistency targets
Jul 16, 2018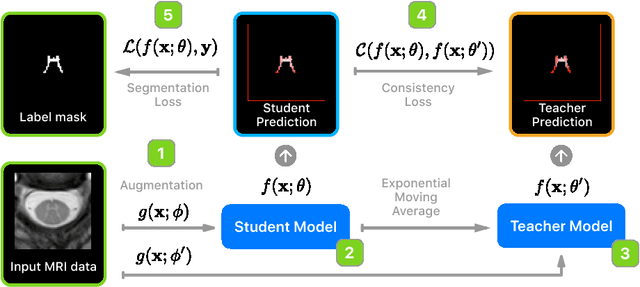
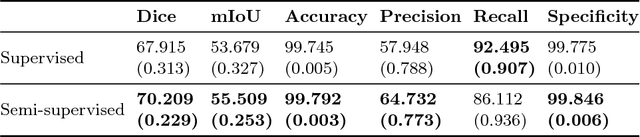
Abstract:Recently proposed techniques for semi-supervised learning such as Temporal Ensembling and Mean Teacher have achieved state-of-the-art results in many important classification benchmarks. In this work, we expand the Mean Teacher approach to segmentation tasks and show that it can bring important improvements in a realistic small data regime using a publicly available multi-center dataset from the Magnetic Resonance Imaging (MRI) domain. We also devise a method to solve the problems that arise when using traditional data augmentation strategies for segmentation tasks on our new training scheme.
AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks
Nov 17, 2017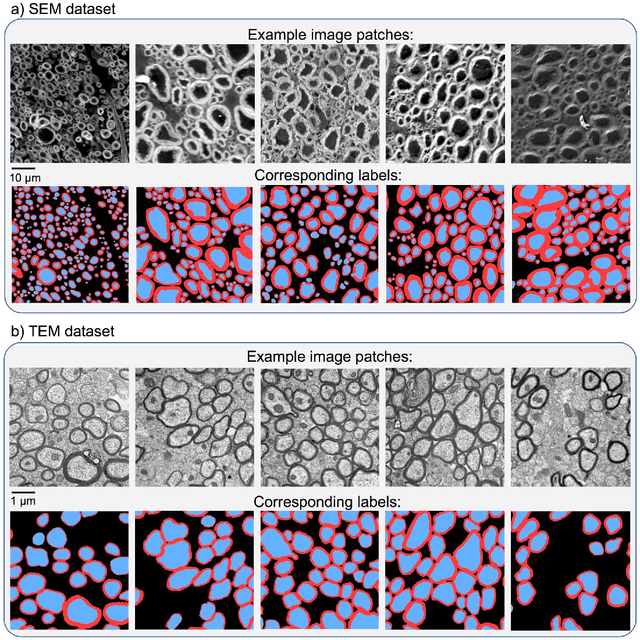
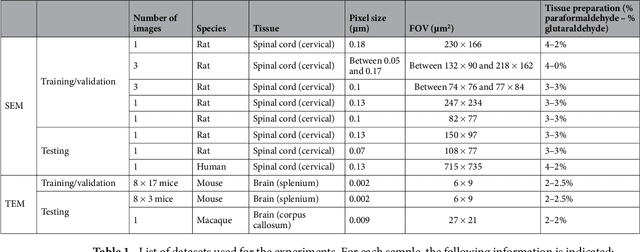
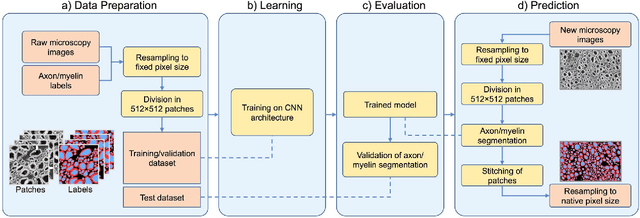
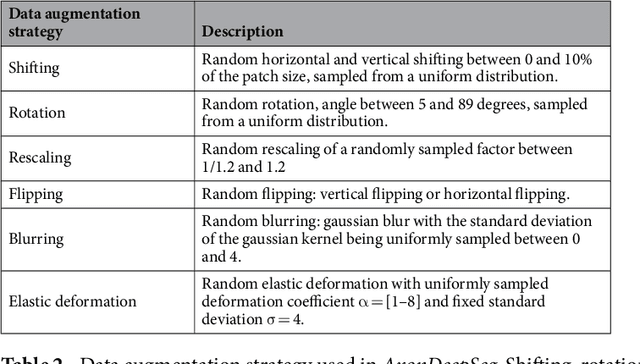
Abstract:Segmentation of axon and myelin from microscopy images of the nervous system provides useful quantitative information about the tissue microstructure, such as axon density and myelin thickness. This could be used for instance to document cell morphometry across species, or to validate novel non-invasive quantitative magnetic resonance imaging techniques. Most currently-available segmentation algorithms are based on standard image processing and usually require multiple processing steps and/or parameter tuning by the user to adapt to different modalities. Moreover, only few methods are publicly available. We introduce AxonDeepSeg, an open-source software that performs axon and myelin segmentation of microscopic images using deep learning. AxonDeepSeg features: (i) a convolutional neural network architecture; (ii) an easy training procedure to generate new models based on manually-labelled data and (iii) two ready-to-use models trained from scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Results show high pixel-wise accuracy across various species: 85% on rat SEM, 81% on human SEM, 95% on mice TEM and 84% on macaque TEM. Segmentation of a full rat spinal cord slice is computed and morphological metrics are extracted and compared against the literature. AxonDeepSeg is freely available at https://github.com/neuropoly/axondeepseg
Spinal cord gray matter segmentation using deep dilated convolutions
Oct 02, 2017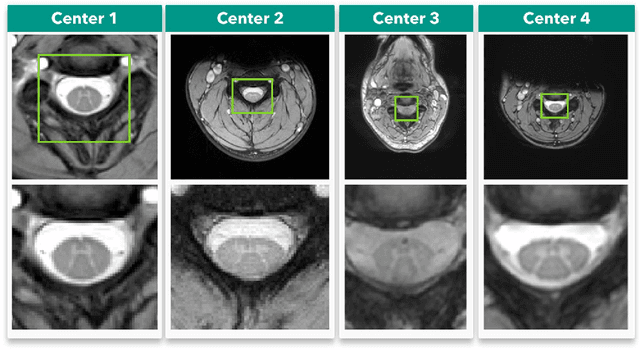
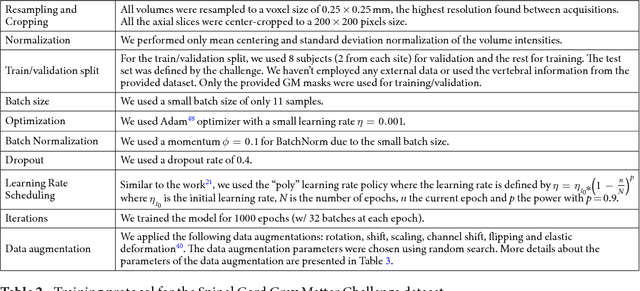
Abstract:Gray matter (GM) tissue changes have been associated with a wide range of neurological disorders and was also recently found relevant as a biomarker for disability in amyotrophic lateral sclerosis. The ability to automatically segment the GM is, therefore, an important task for modern studies of the spinal cord. In this work, we devise a modern, simple and end-to-end fully automated human spinal cord gray matter segmentation method using Deep Learning, that works both on in vivo and ex vivo MRI acquisitions. We evaluate our method against six independently developed methods on a GM segmentation challenge and report state-of-the-art results in 8 out of 10 different evaluation metrics as well as major network parameter reduction when compared to the traditional medical imaging architectures such as U-Nets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge