Jonghye Woo
Memory Consistent Unsupervised Off-the-Shelf Model Adaptation for Source-Relaxed Medical Image Segmentation
Sep 16, 2022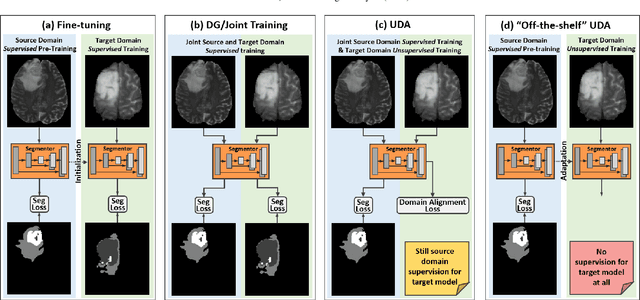
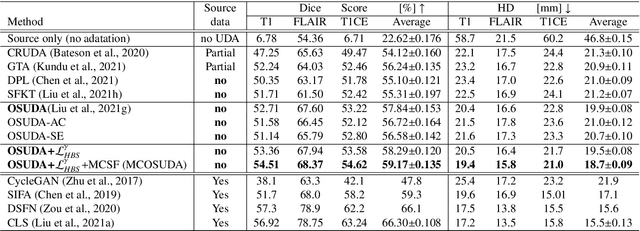
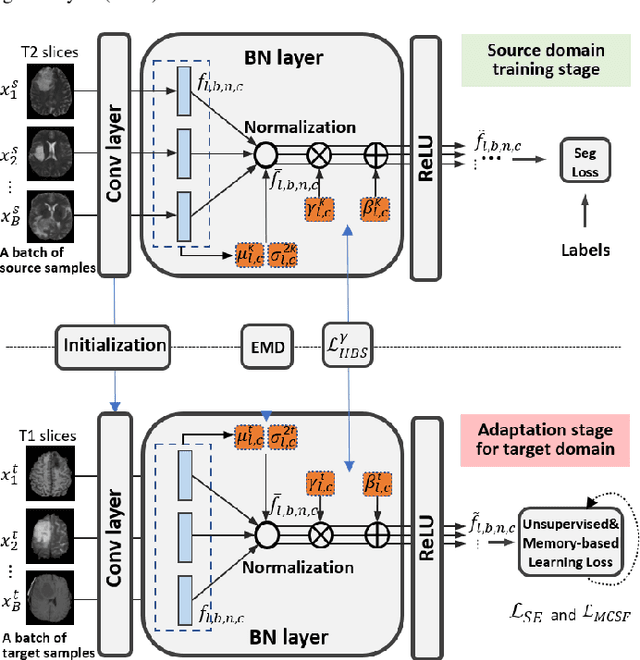
Abstract:Unsupervised domain adaptation (UDA) has been a vital protocol for migrating information learned from a labeled source domain to facilitate the implementation in an unlabeled heterogeneous target domain. Although UDA is typically jointly trained on data from both domains, accessing the labeled source domain data is often restricted, due to concerns over patient data privacy or intellectual property. To sidestep this, we propose "off-the-shelf (OS)" UDA (OSUDA), aimed at image segmentation, by adapting an OS segmentor trained in a source domain to a target domain, in the absence of source domain data in adaptation. Toward this goal, we aim to develop a novel batch-wise normalization (BN) statistics adaptation framework. In particular, we gradually adapt the domain-specific low-order BN statistics, e.g., mean and variance, through an exponential momentum decay strategy, while explicitly enforcing the consistency of the domain shareable high-order BN statistics, e.g., scaling and shifting factors, via our optimization objective. We also adaptively quantify the channel-wise transferability to gauge the importance of each channel, via both low-order statistics divergence and a scaling factor.~Furthermore, we incorporate unsupervised self-entropy minimization into our framework to boost performance alongside a novel queued, memory-consistent self-training strategy to utilize the reliable pseudo label for stable and efficient unsupervised adaptation. We evaluated our OSUDA-based framework on both cross-modality and cross-subtype brain tumor segmentation and cardiac MR to CT segmentation tasks. Our experimental results showed that our memory consistent OSUDA performs better than existing source-relaxed UDA methods and yields similar performance to UDA methods with source data.
Unsupervised Domain Adaptation for Segmentation with Black-box Source Model
Aug 16, 2022Abstract:Unsupervised domain adaptation (UDA) has been widely used to transfer knowledge from a labeled source domain to an unlabeled target domain to counter the difficulty of labeling in a new domain. The training of conventional solutions usually relies on the existence of both source and target domain data. However, privacy of the large-scale and well-labeled data in the source domain and trained model parameters can become the major concern of cross center/domain collaborations. In this work, to address this, we propose a practical solution to UDA for segmentation with a black-box segmentation model trained in the source domain only, rather than original source data or a white-box source model. Specifically, we resort to a knowledge distillation scheme with exponential mixup decay (EMD) to gradually learn target-specific representations. In addition, unsupervised entropy minimization is further applied to regularization of the target domain confidence. We evaluated our framework on the BraTS 2018 database, achieving performance on par with white-box source model adaptation approaches.
Subtype-Aware Dynamic Unsupervised Domain Adaptation
Aug 16, 2022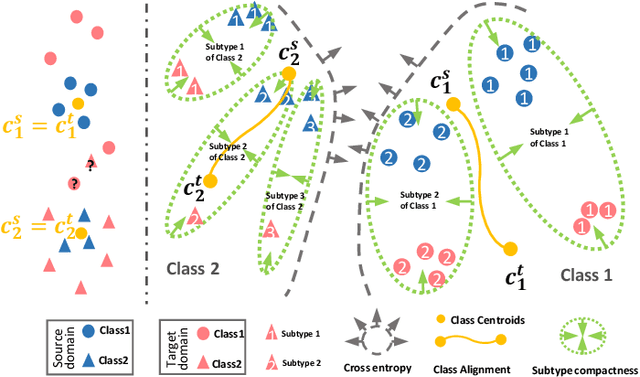
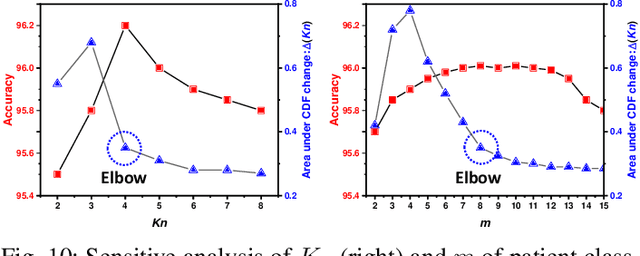
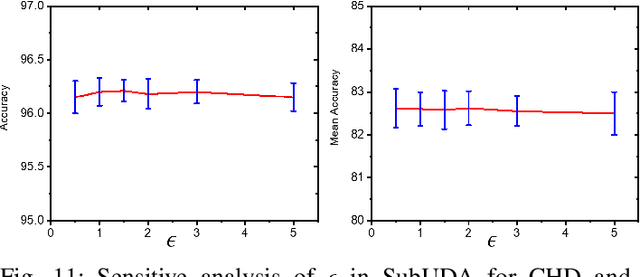
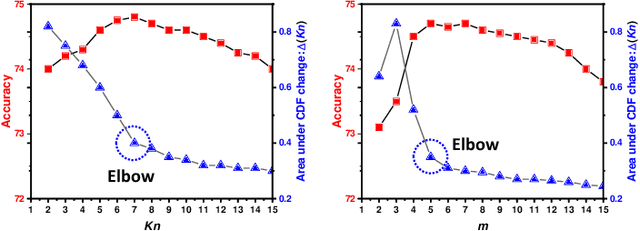
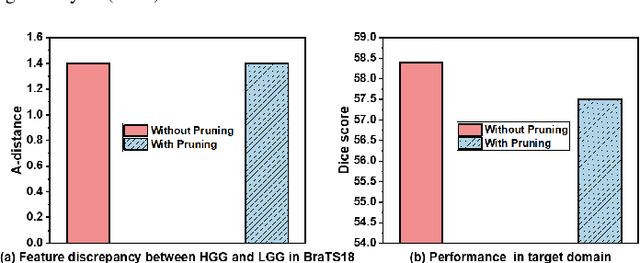
Abstract:Unsupervised domain adaptation (UDA) has been successfully applied to transfer knowledge from a labeled source domain to target domains without their labels. Recently introduced transferable prototypical networks (TPN) further addresses class-wise conditional alignment. In TPN, while the closeness of class centers between source and target domains is explicitly enforced in a latent space, the underlying fine-grained subtype structure and the cross-domain within-class compactness have not been fully investigated. To counter this, we propose a new approach to adaptively perform a fine-grained subtype-aware alignment to improve performance in the target domain without the subtype label in both domains. The insight of our approach is that the unlabeled subtypes in a class have the local proximity within a subtype, while exhibiting disparate characteristics, because of different conditional and label shifts. Specifically, we propose to simultaneously enforce subtype-wise compactness and class-wise separation, by utilizing intermediate pseudo-labels. In addition, we systematically investigate various scenarios with and without prior knowledge of subtype numbers, and propose to exploit the underlying subtype structure. Furthermore, a dynamic queue framework is developed to evolve the subtype cluster centroids steadily using an alternative processing scheme. Experimental results, carried out with multi-view congenital heart disease data and VisDA and DomainNet, show the effectiveness and validity of our subtype-aware UDA, compared with state-of-the-art UDA methods.
Deep Unsupervised Domain Adaptation: A Review of Recent Advances and Perspectives
Aug 15, 2022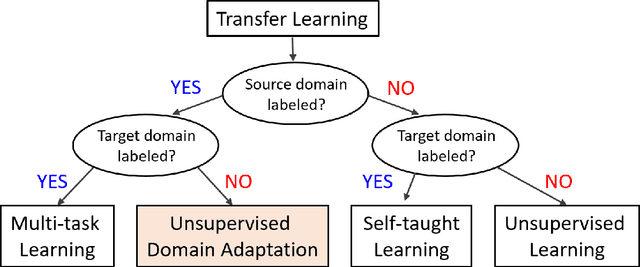

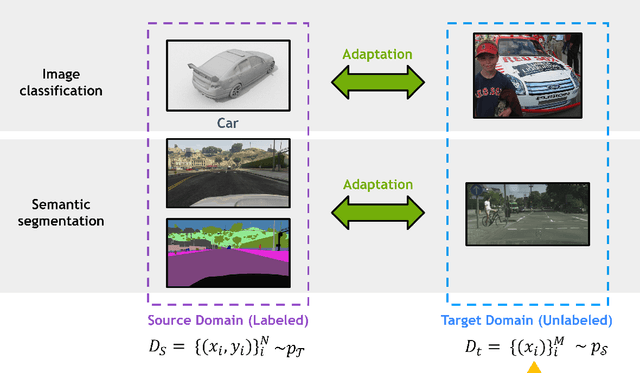
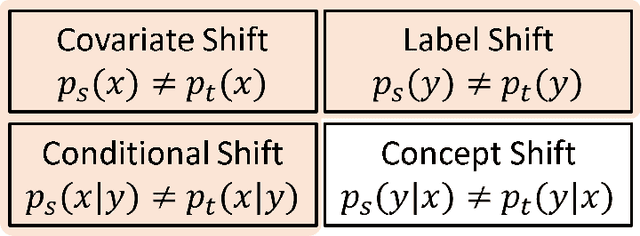
Abstract:Deep learning has become the method of choice to tackle real-world problems in different domains, partly because of its ability to learn from data and achieve impressive performance on a wide range of applications. However, its success usually relies on two assumptions: (i) vast troves of labeled datasets are required for accurate model fitting, and (ii) training and testing data are independent and identically distributed. Its performance on unseen target domains, thus, is not guaranteed, especially when encountering out-of-distribution data at the adaptation stage. The performance drop on data in a target domain is a critical problem in deploying deep neural networks that are successfully trained on data in a source domain. Unsupervised domain adaptation (UDA) is proposed to counter this, by leveraging both labeled source domain data and unlabeled target domain data to carry out various tasks in the target domain. UDA has yielded promising results on natural image processing, video analysis, natural language processing, time-series data analysis, medical image analysis, etc. In this review, as a rapidly evolving topic, we provide a systematic comparison of its methods and applications. In addition, the connection of UDA with its closely related tasks, e.g., domain generalization and out-of-distribution detection, has also been discussed. Furthermore, deficiencies in current methods and possible promising directions are highlighted.
ACT: Semi-supervised Domain-adaptive Medical Image Segmentation with Asymmetric Co-training
Jun 09, 2022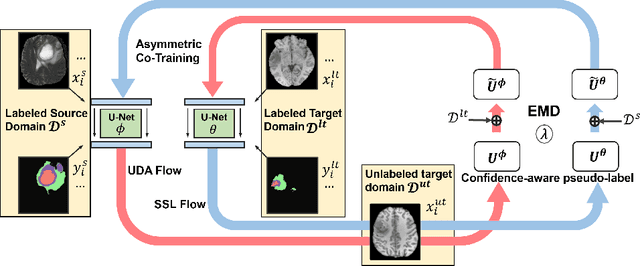
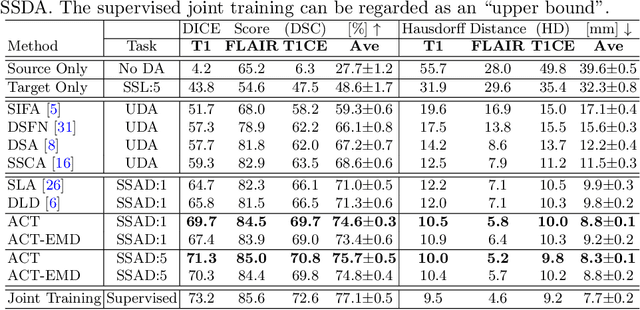
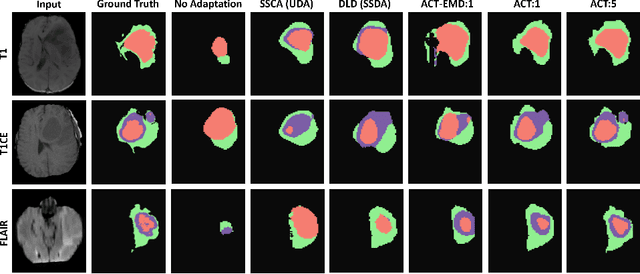
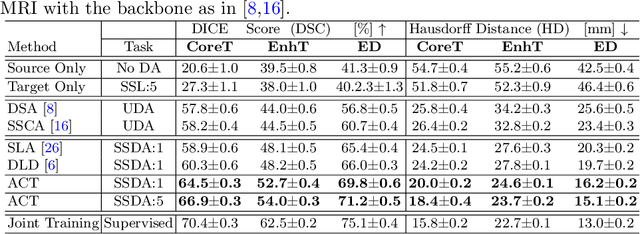
Abstract:Unsupervised domain adaptation (UDA) has been vastly explored to alleviate domain shifts between source and target domains, by applying a well-performed model in an unlabeled target domain via supervision of a labeled source domain. Recent literature, however, has indicated that the performance is still far from satisfactory in the presence of significant domain shifts. Nonetheless, delineating a few target samples is usually manageable and particularly worthwhile, due to the substantial performance gain. Inspired by this, we aim to develop semi-supervised domain adaptation (SSDA) for medical image segmentation, which is largely underexplored. We, thus, propose to exploit both labeled source and target domain data, in addition to unlabeled target data in a unified manner. Specifically, we present a novel asymmetric co-training (ACT) framework to integrate these subsets and avoid the domination of the source domain data. Following a divide-and-conquer strategy, we explicitly decouple the label supervisions in SSDA into two asymmetric sub-tasks, including semi-supervised learning (SSL) and UDA, and leverage different knowledge from two segmentors to take into account the distinction between the source and target label supervisions. The knowledge learned in the two modules is then adaptively integrated with ACT, by iteratively teaching each other, based on the confidence-aware pseudo-label. In addition, pseudo label noise is well-controlled with an exponential MixUp decay scheme for smooth propagation. Experiments on cross-modality brain tumor MRI segmentation tasks using the BraTS18 database showed, even with limited labeled target samples, ACT yielded marked improvements over UDA and state-of-the-art SSDA methods and approached an "upper bound" of supervised joint training.
Tagged-MRI Sequence to Audio Synthesis via Self Residual Attention Guided Heterogeneous Translator
Jun 09, 2022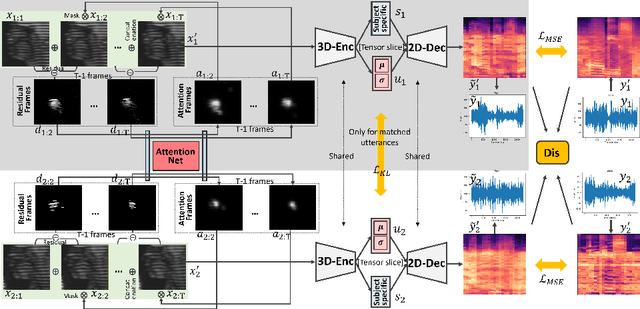
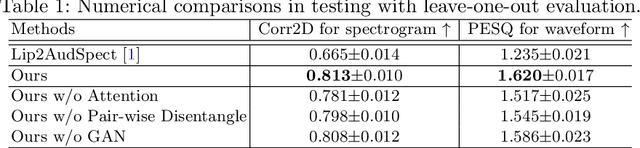
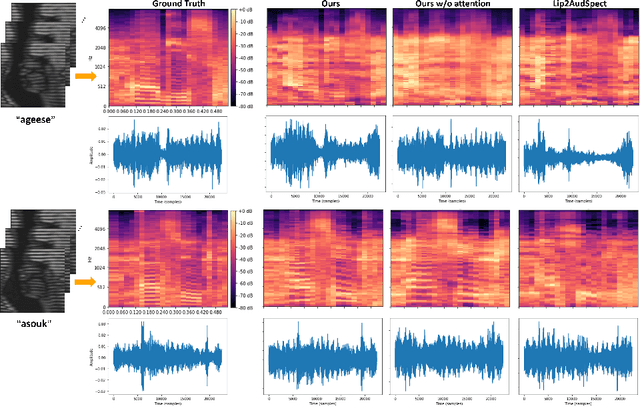
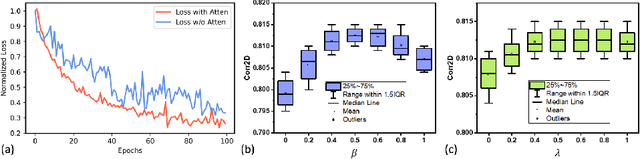
Abstract:Understanding the underlying relationship between tongue and oropharyngeal muscle deformation seen in tagged-MRI and intelligible speech plays an important role in advancing speech motor control theories and treatment of speech related-disorders. Because of their heterogeneous representations, however, direct mapping between the two modalities -- i.e., two-dimensional (mid-sagittal slice) plus time tagged-MRI sequence and its corresponding one-dimensional waveform -- is not straightforward. Instead, we resort to two-dimensional spectrograms as an intermediate representation, which contains both pitch and resonance, from which to develop an end-to-end deep learning framework to translate from a sequence of tagged-MRI to its corresponding audio waveform with limited dataset size.~Our framework is based on a novel fully convolutional asymmetry translator with guidance of a self residual attention strategy to specifically exploit the moving muscular structures during speech.~In addition, we leverage a pairwise correlation of the samples with the same utterances with a latent space representation disentanglement strategy.~Furthermore, we incorporate an adversarial training approach with generative adversarial networks to offer improved realism on our generated spectrograms.~Our experimental results, carried out with a total of 63 tagged-MRI sequences alongside speech acoustics, showed that our framework enabled the generation of clear audio waveforms from a sequence of tagged-MRI, surpassing competing methods. Thus, our framework provides the great potential to help better understand the relationship between the two modalities.
Structure-aware Unsupervised Tagged-to-Cine MRI Synthesis with Self Disentanglement
Feb 25, 2022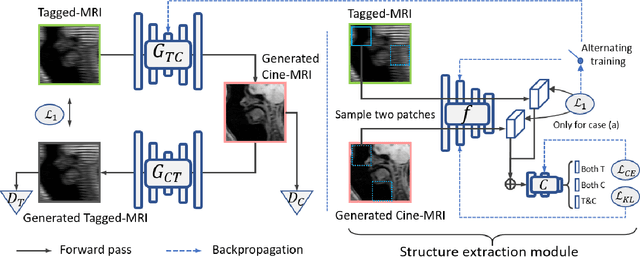
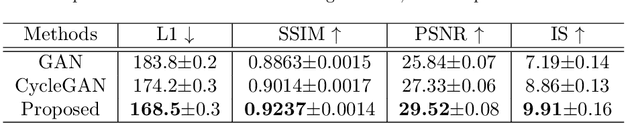
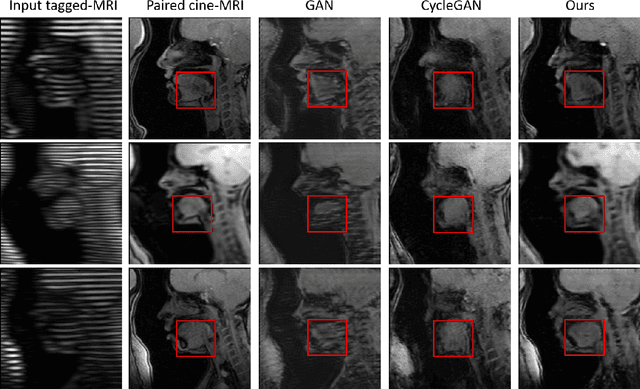
Abstract:Cycle reconstruction regularized adversarial training -- e.g., CycleGAN, DiscoGAN, and DualGAN -- has been widely used for image style transfer with unpaired training data. Several recent works, however, have shown that local distortions are frequent, and structural consistency cannot be guaranteed. Targeting this issue, prior works usually relied on additional segmentation or consistent feature extraction steps that are task-specific. To counter this, this work aims to learn a general add-on structural feature extractor, by explicitly enforcing the structural alignment between an input and its synthesized image. Specifically, we propose a novel input-output image patches self-training scheme to achieve a disentanglement of underlying anatomical structures and imaging modalities. The translator and structure encoder are updated, following an alternating training protocol. In addition, the information w.r.t. imaging modality can be eliminated with an asymmetric adversarial game. We train, validate, and test our network on 1,768, 416, and 1,560 unpaired subject-independent slices of tagged and cine magnetic resonance imaging from a total of twenty healthy subjects, respectively, demonstrating superior performance over competing methods.
Variational Inference for Quantifying Inter-observer Variability in Segmentation of Anatomical Structures
Jan 18, 2022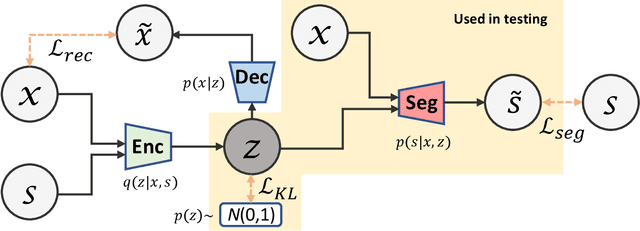

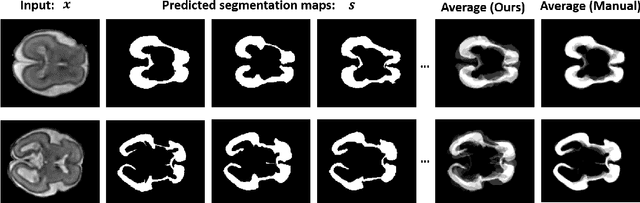

Abstract:Lesions or organ boundaries visible through medical imaging data are often ambiguous, thus resulting in significant variations in multi-reader delineations, i.e., the source of aleatoric uncertainty. In particular, quantifying the inter-observer variability of manual annotations with Magnetic Resonance (MR) Imaging data plays a crucial role in establishing a reference standard for various diagnosis and treatment tasks. Most segmentation methods, however, simply model a mapping from an image to its single segmentation map and do not take the disagreement of annotators into consideration. In order to account for inter-observer variability, without sacrificing accuracy, we propose a novel variational inference framework to model the distribution of plausible segmentation maps, given a specific MR image, which explicitly represents the multi-reader variability. Specifically, we resort to a latent vector to encode the multi-reader variability and counteract the inherent information loss in the imaging data. Then, we apply a variational autoencoder network and optimize its evidence lower bound (ELBO) to efficiently approximate the distribution of the segmentation map, given an MR image. Experimental results, carried out with the QUBIQ brain growth MRI segmentation datasets with seven annotators, demonstrate the effectiveness of our approach.
Self-semantic contour adaptation for cross modality brain tumor segmentation
Jan 13, 2022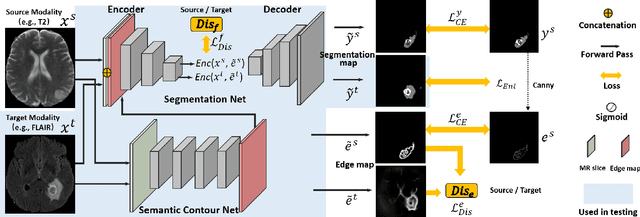
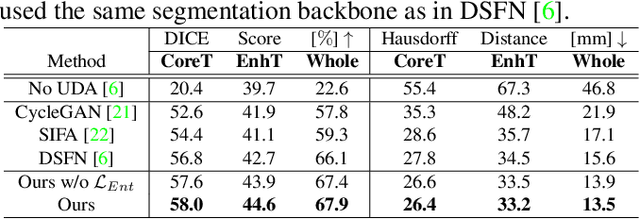
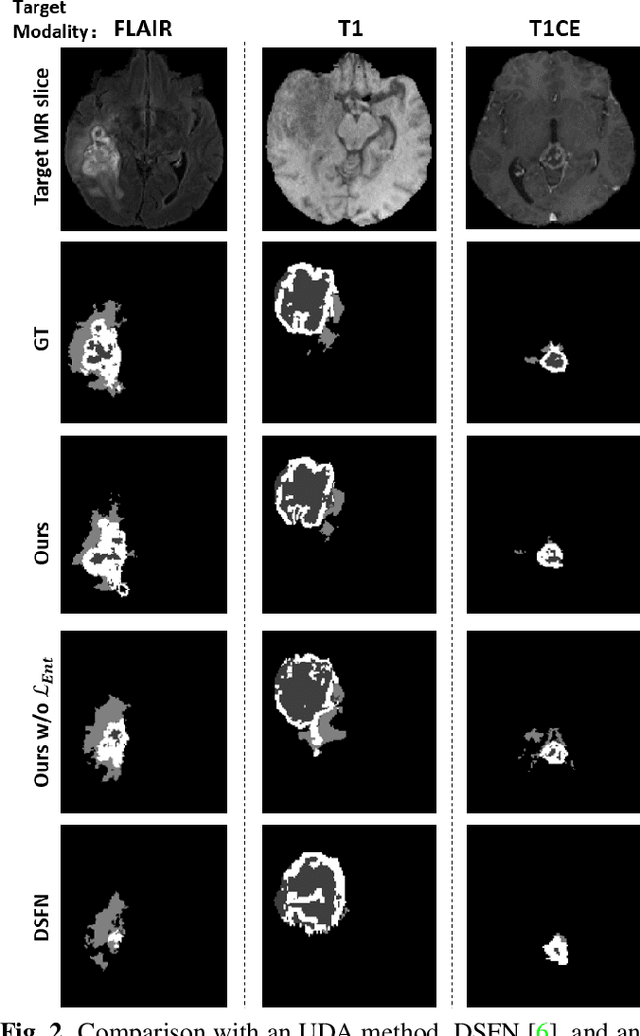
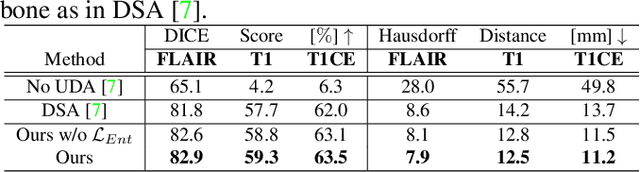
Abstract:Unsupervised domain adaptation (UDA) between two significantly disparate domains to learn high-level semantic alignment is a crucial yet challenging task.~To this end, in this work, we propose exploiting low-level edge information to facilitate the adaptation as a precursor task, which has a small cross-domain gap, compared with semantic segmentation.~The precise contour then provides spatial information to guide the semantic adaptation. More specifically, we propose a multi-task framework to learn a contouring adaptation network along with a semantic segmentation adaptation network, which takes both magnetic resonance imaging (MRI) slice and its initial edge map as input.~These two networks are jointly trained with source domain labels, and the feature and edge map level adversarial learning is carried out for cross-domain alignment. In addition, self-entropy minimization is incorporated to further enhance segmentation performance. We evaluated our framework on the BraTS2018 database for cross-modality segmentation of brain tumors, showing the validity and superiority of our approach, compared with competing methods.
Adversarial Unsupervised Domain Adaptation with Conditional and Label Shift: Infer, Align and Iterate
Aug 02, 2021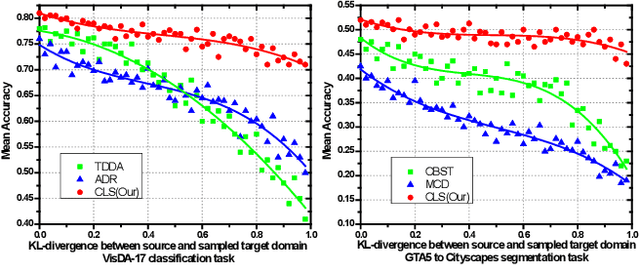
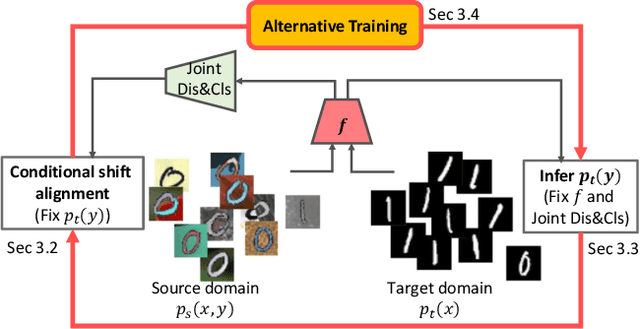
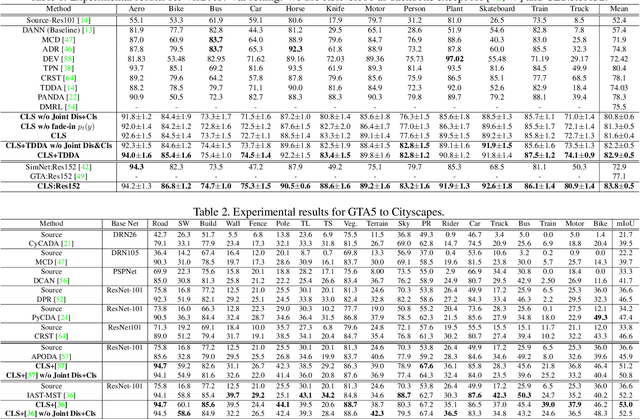
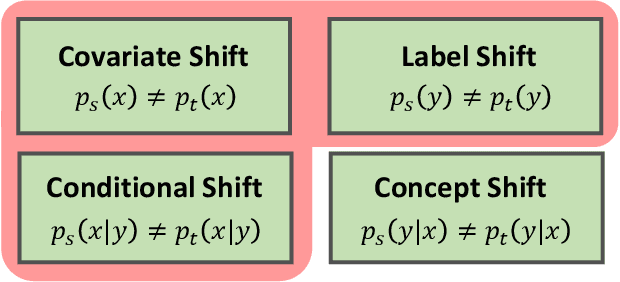
Abstract:In this work, we propose an adversarial unsupervised domain adaptation (UDA) approach with the inherent conditional and label shifts, in which we aim to align the distributions w.r.t. both $p(x|y)$ and $p(y)$. Since the label is inaccessible in the target domain, the conventional adversarial UDA assumes $p(y)$ is invariant across domains, and relies on aligning $p(x)$ as an alternative to the $p(x|y)$ alignment. To address this, we provide a thorough theoretical and empirical analysis of the conventional adversarial UDA methods under both conditional and label shifts, and propose a novel and practical alternative optimization scheme for adversarial UDA. Specifically, we infer the marginal $p(y)$ and align $p(x|y)$ iteratively in the training, and precisely align the posterior $p(y|x)$ in testing. Our experimental results demonstrate its effectiveness on both classification and segmentation UDA, and partial UDA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge