Jie Hua
Benchmarking Preprocessing and Integration Methods in Single-Cell Genomics
Jan 01, 2026Abstract:Single-cell data analysis has the potential to revolutionize personalized medicine by characterizing disease-associated molecular changes at the single-cell level. Advanced single-cell multimodal assays can now simultaneously measure various molecules (e.g., DNA, RNA, Protein) across hundreds of thousands of individual cells, providing a comprehensive molecular readout. A significant analytical challenge is integrating single-cell measurements across different modalities. Various methods have been developed to address this challenge, but there has been no systematic evaluation of these techniques with different preprocessing strategies. This study examines a general pipeline for single-cell data analysis, which includes normalization, data integration, and dimensionality reduction. The performance of different algorithm combinations often depends on the dataset sizes and characteristics. We evaluate six datasets across diverse modalities, tissues, and organisms using three metrics: Silhouette Coefficient Score, Adjusted Rand Index, and Calinski-Harabasz Index. Our experiments involve combinations of seven normalization methods, four dimensional reduction methods, and five integration methods. The results show that Seurat and Harmony excel in data integration, with Harmony being more time-efficient, especially for large datasets. UMAP is the most compatible dimensionality reduction method with the integration techniques, and the choice of normalization method varies depending on the integration method used.
Towards Automated Differential Diagnosis of Skin Diseases Using Deep Learning and Imbalance-Aware Strategies
Jan 01, 2026Abstract:As dermatological conditions become increasingly common and the availability of dermatologists remains limited, there is a growing need for intelligent tools to support both patients and clinicians in the timely and accurate diagnosis of skin diseases. In this project, we developed a deep learning based model for the classification and diagnosis of skin conditions. By leveraging pretraining on publicly available skin disease image datasets, our model effectively extracted visual features and accurately classified various dermatological cases. Throughout the project, we refined the model architecture, optimized data preprocessing workflows, and applied targeted data augmentation techniques to improve overall performance. The final model, based on the Swin Transformer, achieved a prediction accuracy of 87.71 percent across eight skin lesion classes on the ISIC2019 dataset. These results demonstrate the model's potential as a diagnostic support tool for clinicians and a self assessment aid for patients.
Opportunistic Federated Learning: An Exploration of Egocentric Collaboration for Pervasive Computing Applications
Mar 24, 2021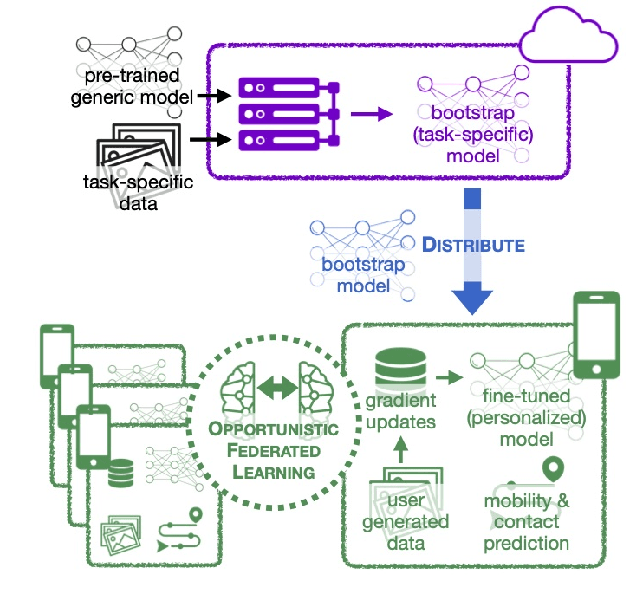
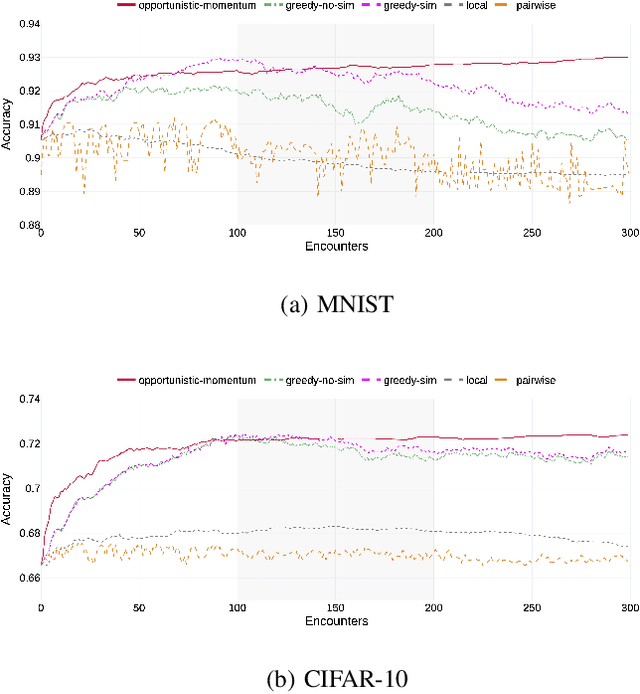
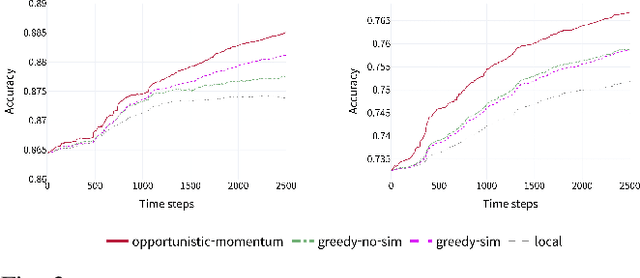
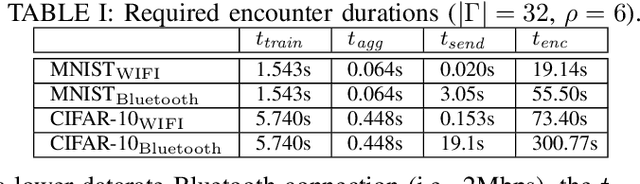
Abstract:Pervasive computing applications commonly involve user's personal smartphones collecting data to influence application behavior. Applications are often backed by models that learn from the user's experiences to provide personalized and responsive behavior. While models are often pre-trained on massive datasets, federated learning has gained attention for its ability to train globally shared models on users' private data without requiring the users to share their data directly. However, federated learning requires devices to collaborate via a central server, under the assumption that all users desire to learn the same model. We define a new approach, opportunistic federated learning, in which individual devices belonging to different users seek to learn robust models that are personalized to their user's own experiences. However, instead of learning in isolation, these models opportunistically incorporate the learned experiences of other devices they encounter opportunistically. In this paper, we explore the feasibility and limits of such an approach, culminating in a framework that supports encounter-based pairwise collaborative learning. The use of our opportunistic encounter-based learning amplifies the performance of personalized learning while resisting overfitting to encountered data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge