Jianning Li
Open-Source Skull Reconstruction with MONAI
Nov 25, 2022



Abstract:We present a deep learning-based approach for skull reconstruction for MONAI, which has been pre-trained on the MUG500+ skull dataset. The implementation follows the MONAI contribution guidelines, hence, it can be easily tried out and used, and extended by MONAI users. The primary goal of this paper lies in the investigation of open-sourcing codes and pre-trained deep learning models under the MONAI framework. Nowadays, open-sourcing software, especially (pre-trained) deep learning models, has become increasingly important. Over the years, medical image analysis experienced a tremendous transformation. Over a decade ago, algorithms had to be implemented and optimized with low-level programming languages, like C or C++, to run in a reasonable time on a desktop PC, which was not as powerful as today's computers. Nowadays, users have high-level scripting languages like Python, and frameworks like PyTorch and TensorFlow, along with a sea of public code repositories at hand. As a result, implementations that had thousands of lines of C or C++ code in the past, can now be scripted with a few lines and in addition executed in a fraction of the time. To put this even on a higher level, the Medical Open Network for Artificial Intelligence (MONAI) framework tailors medical imaging research to an even more convenient process, which can boost and push the whole field. The MONAI framework is a freely available, community-supported, open-source and PyTorch-based framework, that also enables to provide research contributions with pre-trained models to others. Codes and pre-trained weights for skull reconstruction are publicly available at: https://github.com/Project-MONAI/research-contributions/tree/master/SkullRec
Training β-VAE by Aggregating a Learned Gaussian Posterior with a Decoupled Decoder
Sep 29, 2022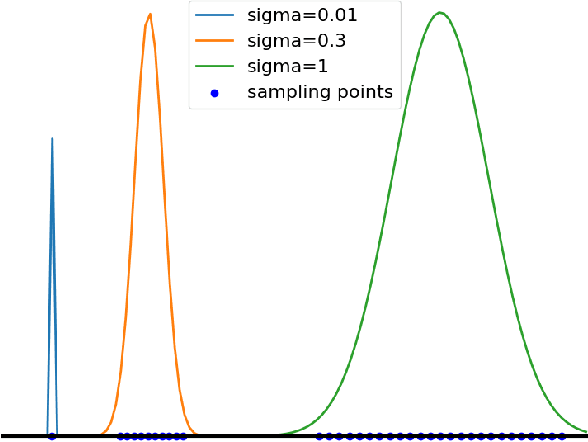
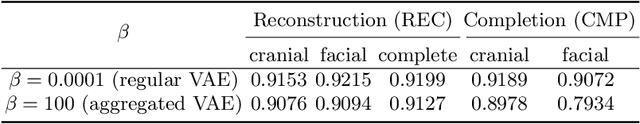
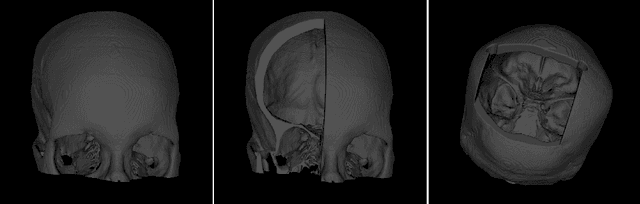
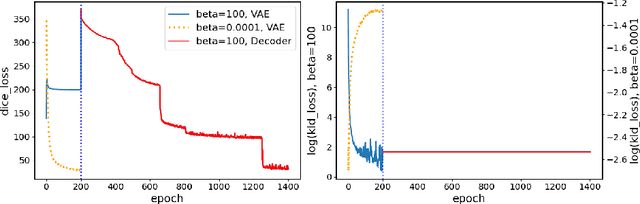
Abstract:The reconstruction loss and the Kullback-Leibler divergence (KLD) loss in a variational autoencoder (VAE) often play antagonistic roles, and tuning the weight of the KLD loss in $\beta$-VAE to achieve a balance between the two losses is a tricky and dataset-specific task. As a result, current practices in VAE training often result in a trade-off between the reconstruction fidelity and the continuity$/$disentanglement of the latent space, if the weight $\beta$ is not carefully tuned. In this paper, we present intuitions and a careful analysis of the antagonistic mechanism of the two losses, and propose, based on the insights, a simple yet effective two-stage method for training a VAE. Specifically, the method aggregates a learned Gaussian posterior $z \sim q_{\theta} (z|x)$ with a decoder decoupled from the KLD loss, which is trained to learn a new conditional distribution $p_{\phi} (x|z)$ of the input data $x$. Experimentally, we show that the aggregated VAE maximally satisfies the Gaussian assumption about the latent space, while still achieves a reconstruction error comparable to when the latent space is only loosely regularized by $\mathcal{N}(\mathbf{0},I)$. The proposed approach does not require hyperparameter (i.e., the KLD weight $\beta$) tuning given a specific dataset as required in common VAE training practices. We evaluate the method using a medical dataset intended for 3D skull reconstruction and shape completion, and the results indicate promising generative capabilities of the VAE trained using the proposed method. Besides, through guided manipulation of the latent variables, we establish a connection between existing autoencoder (AE)-based approaches and generative approaches, such as VAE, for the shape completion problem. Codes and pre-trained weights are available at https://github.com/Jianningli/skullVAE
The HoloLens in Medicine: A systematic Review and Taxonomy
Sep 06, 2022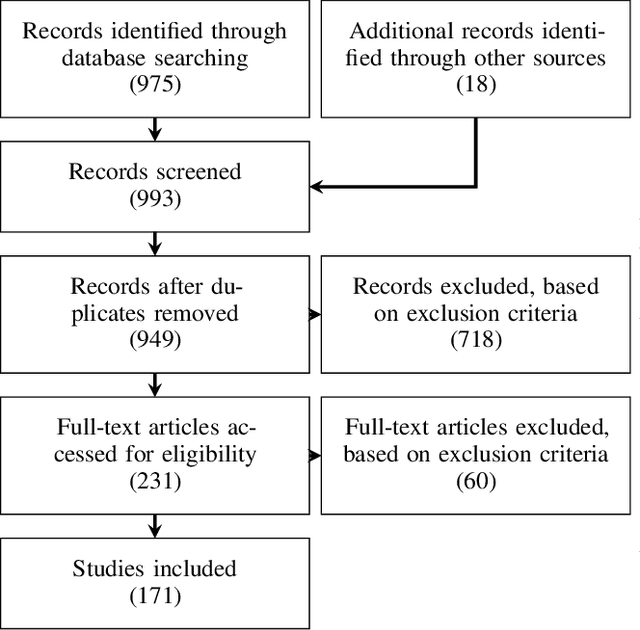
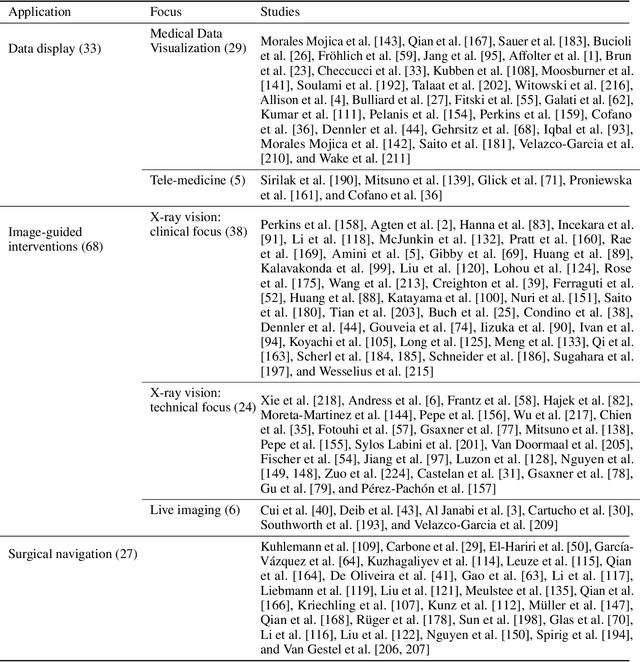
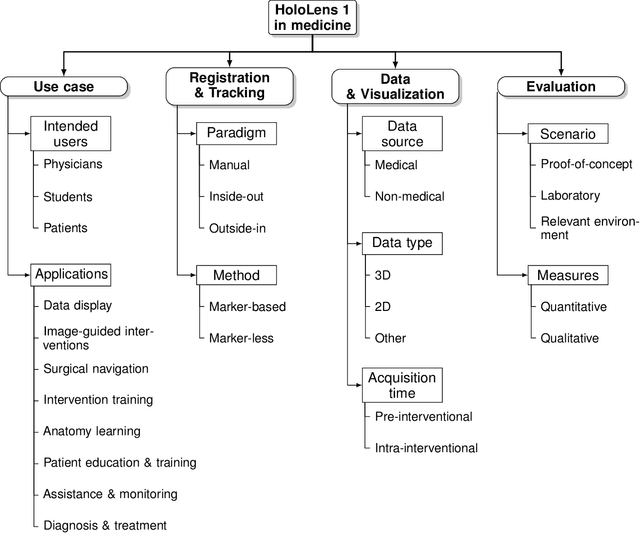

Abstract:The HoloLens (Microsoft Corp., Redmond, WA), a head-worn, optically see-through augmented reality display, is the main player in the recent boost in medical augmented reality research. In medical settings, the HoloLens enables the physician to obtain immediate insight into patient information, directly overlaid with their view of the clinical scenario, the medical student to gain a better understanding of complex anatomies or procedures, and even the patient to execute therapeutic tasks with improved, immersive guidance. In this systematic review, we provide a comprehensive overview of the usage of the first-generation HoloLens within the medical domain, from its release in March 2016, until the year of 2021, were attention is shifting towards it's successor, the HoloLens 2. We identified 171 relevant publications through a systematic search of the PubMed and Scopus databases. We analyze these publications in regard to their intended use case, technical methodology for registration and tracking, data sources, visualization as well as validation and evaluation. We find that, although the feasibility of using the HoloLens in various medical scenarios has been shown, increased efforts in the areas of precision, reliability, usability, workflow and perception are necessary to establish AR in clinical practice.
GAN-based generation of realistic 3D data: A systematic review and taxonomy
Jul 04, 2022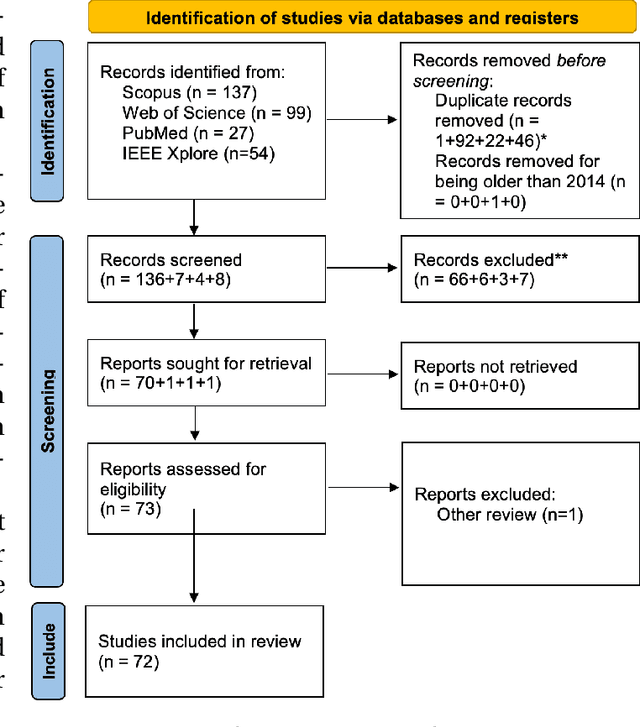
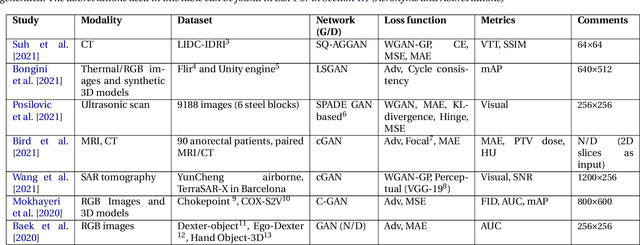
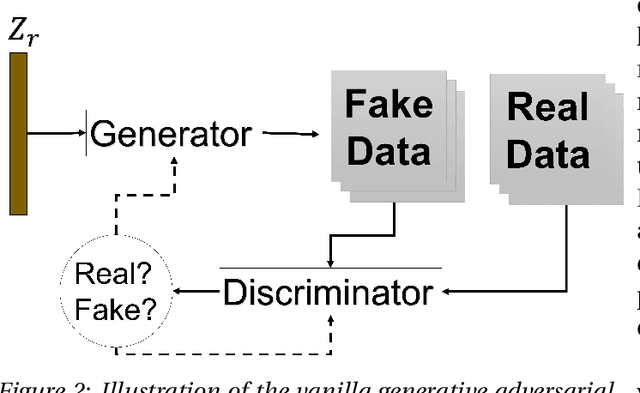
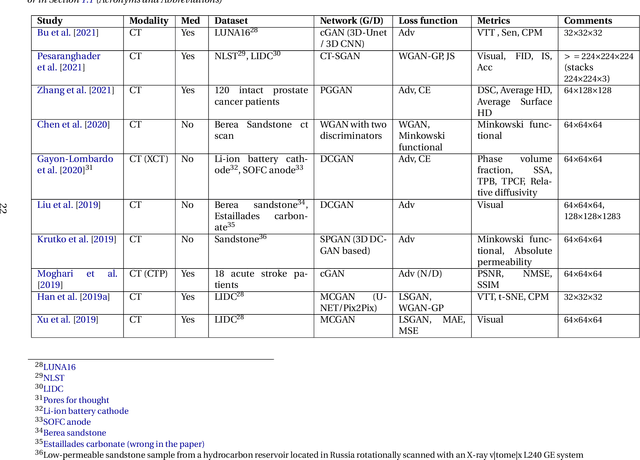
Abstract:Data has become the most valuable resource in today's world. With the massive proliferation of data-driven algorithms, such as deep learning-based approaches, the availability of data is of great interest. In this context, high-quality training, validation and testing datasets are particularly needed. Volumetric data is a very important resource in medicine, as it ranges from disease diagnoses to therapy monitoring. When the dataset is sufficient, models can be trained to help doctors with these tasks. Unfortunately, there are scenarios and applications where large amounts of data is unavailable. For example, in the medical field, rare diseases and privacy issues can lead to restricted data availability. In non-medical fields, the high cost of obtaining a sufficient amount of high-quality data can also be a concern. A solution to these problems can be the generation of synthetic data to perform data augmentation in combination with other more traditional methods of data augmentation. Therefore, most of the publications on 3D Generative Adversarial Networks (GANs) are within the medical domain. The existence of mechanisms to generate realistic synthetic data is a good asset to overcome this challenge, especially in healthcare, as the data must be of good quality and close to reality, i.e. realistic, and without privacy issues. In this review, we provide a summary of works that generate realistic 3D synthetic data using GANs. We therefore outline GAN-based methods in these areas with common architectures, advantages and disadvantages. We present a novel taxonomy, evaluations, challenges and research opportunities to provide a holistic overview of the current state of GANs in medicine and other fields.
Back to the Roots: Reconstructing Large and Complex Cranial Defects using an Image-based Statistical Shape Model
Apr 12, 2022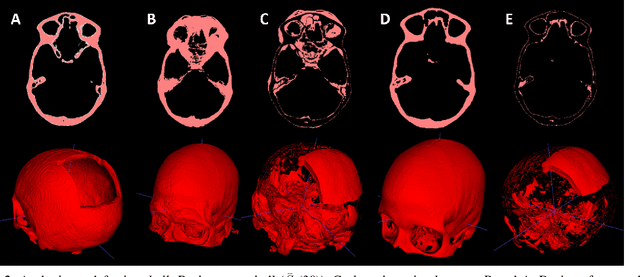
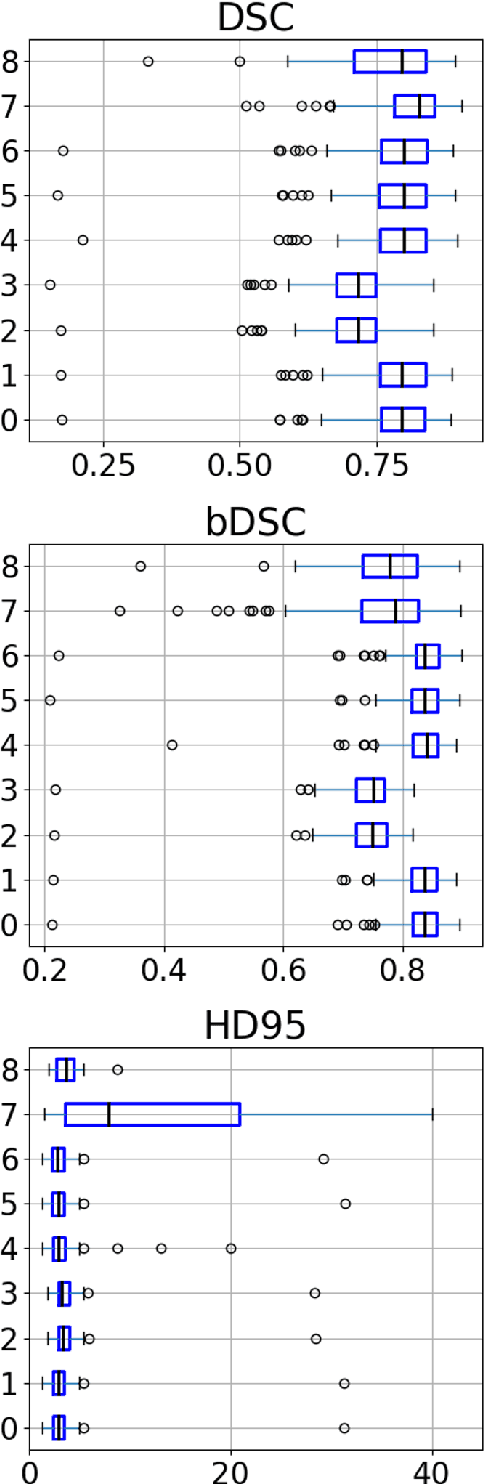
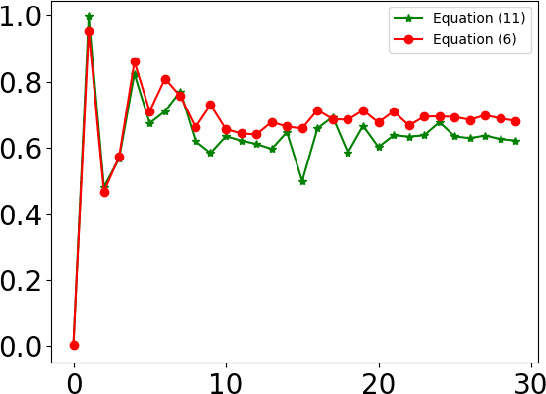
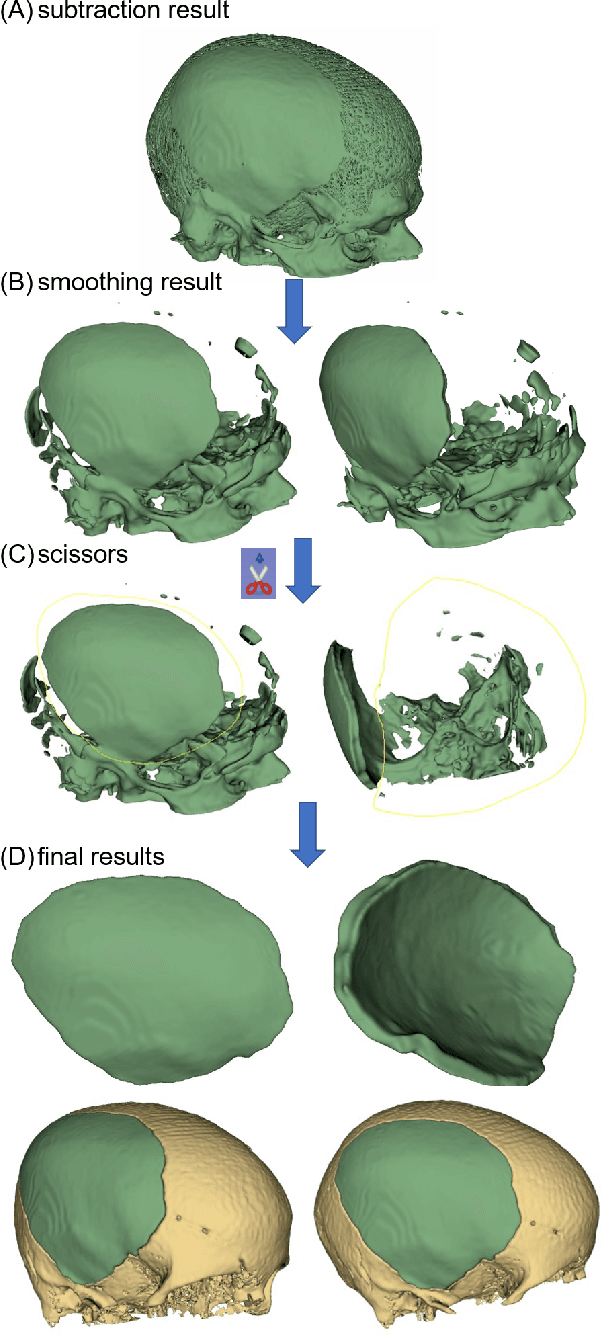
Abstract:Designing implants for large and complex cranial defects is a challenging task, even for professional designers. Current efforts on automating the design process focused mainly on convolutional neural networks (CNN), which have produced state-of-the-art results on reconstructing synthetic defects. However, existing CNN-based methods have been difficult to translate to clinical practice in cranioplasty, as their performance on complex and irregular cranial defects remains unsatisfactory. In this paper, a statistical shape model (SSM) built directly on the segmentation masks of the skulls is presented. We evaluate the SSM on several cranial implant design tasks, and the results show that, while the SSM performs suboptimally on synthetic defects compared to CNN-based approaches, it is capable of reconstructing large and complex defects with only minor manual corrections. The quality of the resulting implants is examined and assured by experienced neurosurgeons. In contrast, CNN-based approaches, even with massive data augmentation, fail or produce less-than-satisfactory implants for these cases. Codes are publicly available at https://github.com/Jianningli/ssm
Automated cross-sectional view selection in CT angiography of aortic dissections with uncertainty awareness and retrospective clinical annotations
Nov 22, 2021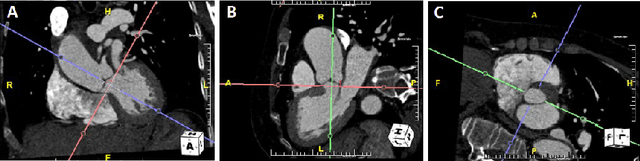
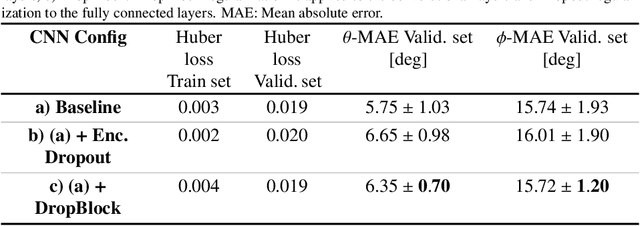
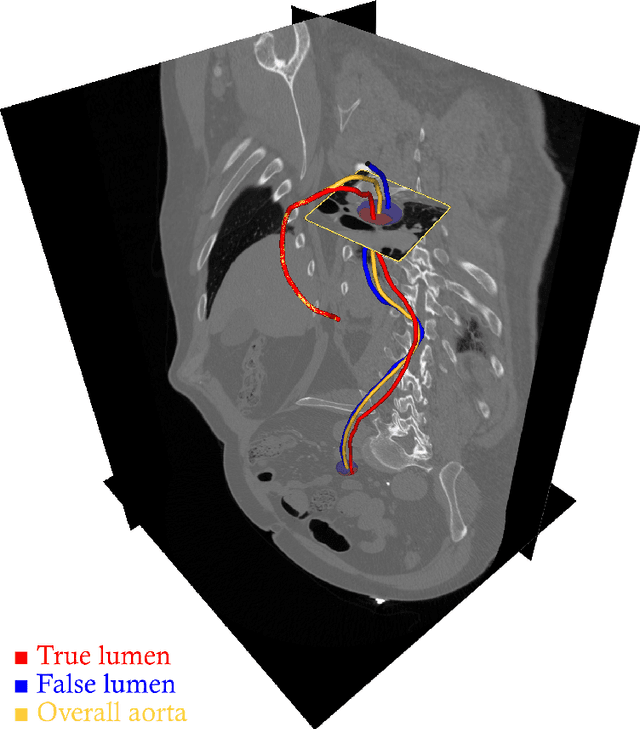
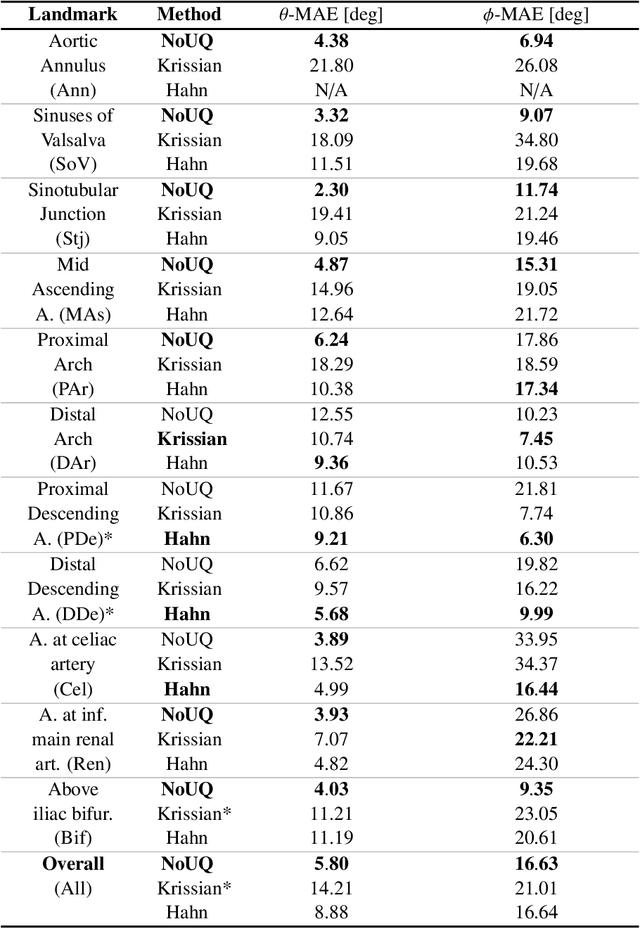
Abstract:Objective: Surveillance imaging of chronic aortic diseases, such as dissections, relies on obtaining and comparing cross-sectional diameter measurements at predefined aortic landmarks, over time. Due to a lack of robust tools, the orientation of the cross-sectional planes is defined manually by highly trained operators. We show how manual annotations routinely collected in a clinic can be efficiently used to ease this task, despite the presence of a non-negligible interoperator variability in the measurements. Impact: Ill-posed but repetitive imaging tasks can be eased or automated by leveraging imperfect, retrospective clinical annotations. Methodology: In this work, we combine convolutional neural networks and uncertainty quantification methods to predict the orientation of such cross-sectional planes. We use clinical data randomly processed by 11 operators for training, and test on a smaller set processed by 3 independent operators to assess interoperator variability. Results: Our analysis shows that manual selection of cross-sectional planes is characterized by 95% limits of agreement (LOA) of $10.6^\circ$ and $21.4^\circ$ per angle. Our method showed to decrease static error by $3.57^\circ$ ($40.2$%) and $4.11^\circ$ ($32.8$%) against state of the art and LOA by $5.4^\circ$ ($49.0$%) and $16.0^\circ$ ($74.6$%) against manual processing. Conclusion: This suggests that pre-existing annotations can be an inexpensive resource in clinics to ease ill-posed and repetitive tasks like cross-section extraction for surveillance of aortic dissections.
Learning to Rearrange Voxels in Binary Segmentation Masks for Smooth Manifold Triangulation
Aug 11, 2021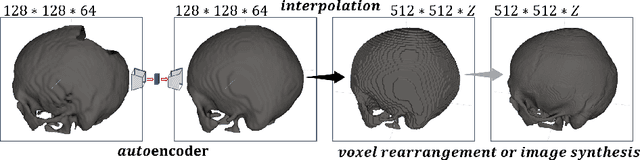
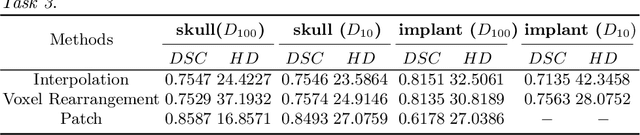
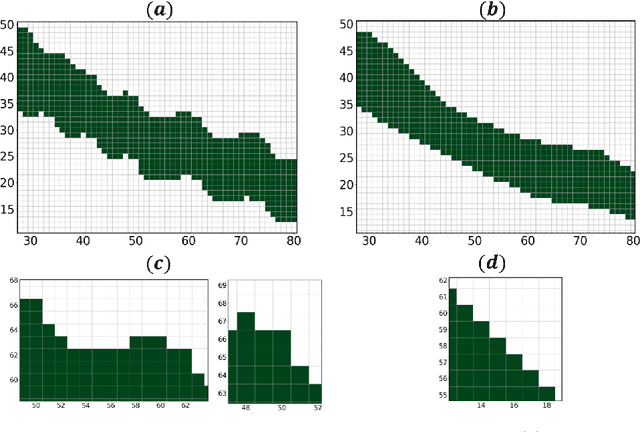
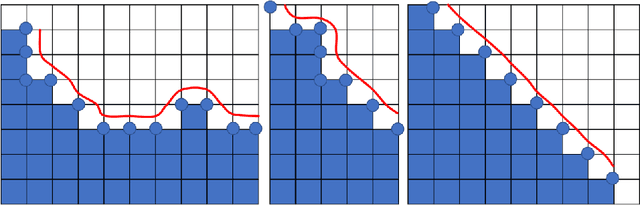
Abstract:Medical images, especially volumetric images, are of high resolution and often exceed the capacity of standard desktop GPUs. As a result, most deep learning-based medical image analysis tasks require the input images to be downsampled, often substantially, before these can be fed to a neural network. However, downsampling can lead to a loss of image quality, which is undesirable especially in reconstruction tasks, where the fine geometric details need to be preserved. In this paper, we propose that high-resolution images can be reconstructed in a coarse-to-fine fashion, where a deep learning algorithm is only responsible for generating a coarse representation of the image, which consumes moderate GPU memory. For producing the high-resolution outcome, we propose two novel methods: learned voxel rearrangement of the coarse output and hierarchical image synthesis. Compared to the coarse output, the high-resolution counterpart allows for smooth surface triangulation, which can be 3D-printed in the highest possible quality. Experiments of this paper are carried out on the dataset of AutoImplant 2021 (https://autoimplant2021.grand-challenge.org/), a MICCAI challenge on cranial implant design. The dataset contains high-resolution skulls that can be viewed as 2D manifolds embedded in a 3D space. Codes associated with this study can be accessed at https://github.com/Jianningli/voxel_rearrangement.
AI-based Aortic Vessel Tree Segmentation for Cardiovascular Diseases Treatment: Status Quo
Aug 06, 2021
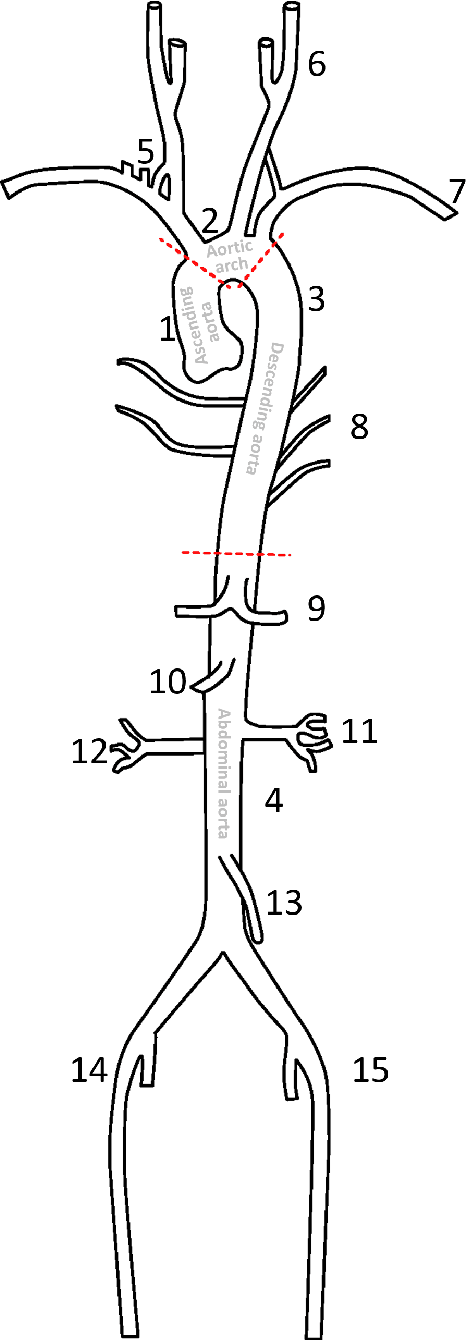
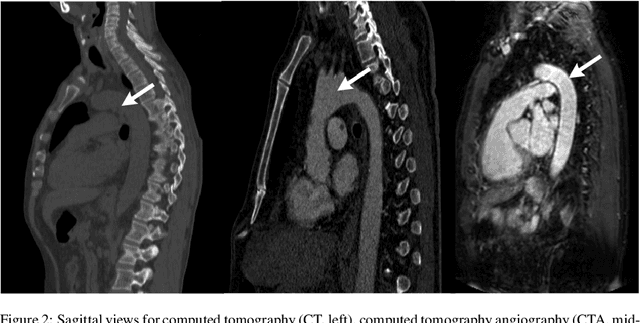
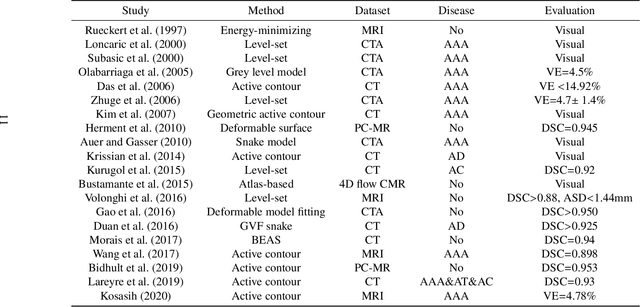
Abstract:The aortic vessel tree is composed of the aorta and its branching arteries, and plays a key role in supplying the whole body with blood. Aortic diseases, like aneurysms or dissections, can lead to an aortic rupture, whose treatment with open surgery is highly risky. Therefore, patients commonly undergo drug treatment under constant monitoring, which requires regular inspections of the vessels through imaging. The standard imaging modality for diagnosis and monitoring is computed tomography (CT), which can provide a detailed picture of the aorta and its branching vessels if combined with a contrast agent, resulting in a CT angiography (CTA). Optimally, the whole aortic vessel tree geometry from consecutive CTAs, are overlaid and compared. This allows to not only detect changes in the aorta, but also more peripheral vessel tree changes, caused by the primary pathology or newly developed. When performed manually, this reconstruction requires slice by slice contouring, which could easily take a whole day for a single aortic vessel tree and, hence, is not feasible in clinical practice. Automatic or semi-automatic vessel tree segmentation algorithms, on the other hand, can complete this task in a fraction of the manual execution time and run in parallel to the clinical routine of the clinicians. In this paper, we systematically review computing techniques for the automatic and semi-automatic segmentation of the aortic vessel tree. The review concludes with an in-depth discussion on how close these state-of-the-art approaches are to an application in clinical practice and how active this research field is, taking into account the number of publications, datasets and challenges.
Deep Learning -- A first Meta-Survey of selected Reviews across Scientific Disciplines and their Research Impact
Nov 16, 2020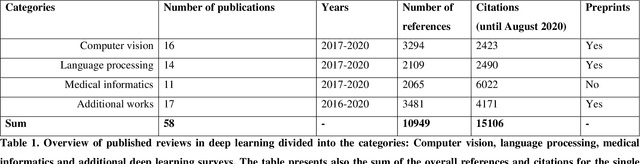
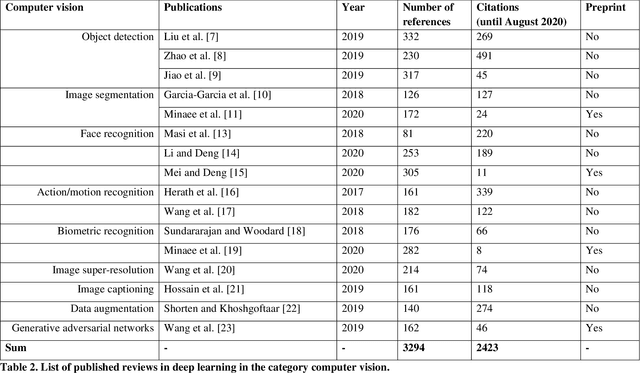
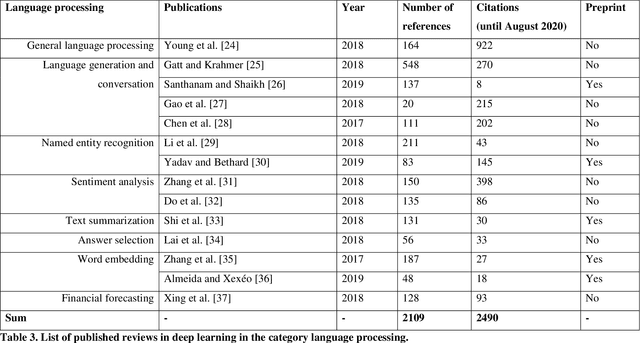
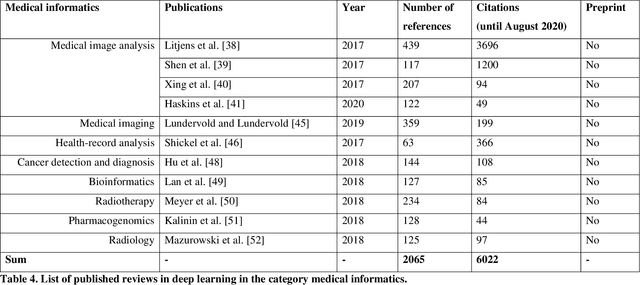
Abstract:Deep learning belongs to the field of artificial intelligence, where machines perform tasks that typically require some kind of human intelligence. Deep learning tries to achieve this by mimicking the learning of a human brain. Similar to the basic structure of a brain, which consists of (billions of) neurons and connections between them, a deep learning algorithm consists of an artificial neural network, which resembles the biological brain structure. Mimicking the learning process of humans with their senses, deep learning networks are fed with (sensory) data, like texts, images, videos or sounds. These networks outperform the state-of-the-art methods in different tasks and, because of this, the whole field saw an exponential growth during the last years. This growth resulted in way over 10 000 publications per year in the last years. For example, the search engine PubMed alone, which covers only a sub-set of all publications in the medical field, provides over 11 000 results for the search term $'$deep learning$'$ in Q3 2020, and ~90% of these results are from the last three years. Consequently, a complete overview over the field of deep learning is already impossible to obtain and, in the near future, it will potentially become difficult to obtain an overview over a subfield. However, there are several review articles about deep learning, which are focused on specific scientific fields or applications, for example deep learning advances in computer vision or in specific tasks like object detection. With these surveys as a foundation, the aim of this contribution is to provide a first high-level, categorized meta-analysis of selected reviews on deep learning across different scientific disciplines and outline the research impact that they already have during a short period of time.
Medical Deep Learning -- A systematic Meta-Review
Oct 28, 2020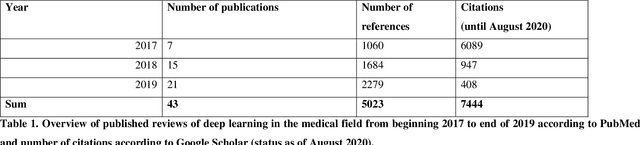
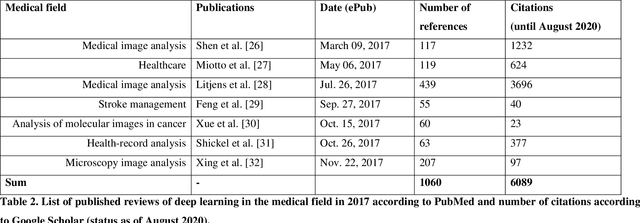
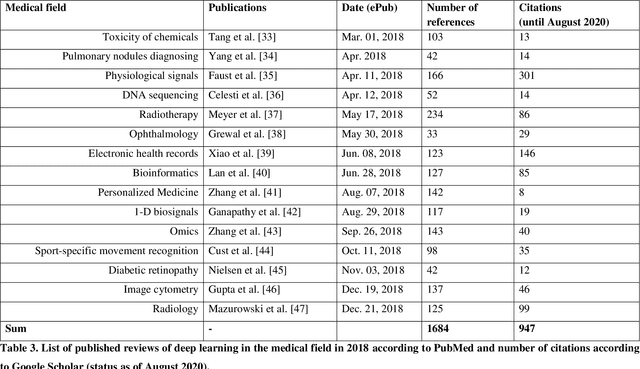

Abstract:Deep learning had a remarkable impact in different scientific disciplines during the last years. This was demonstrated in numerous tasks, where deep learning algorithms were able to outperform the state-of-art methods, also in image processing and analysis. Moreover, deep learning delivers good results in tasks like autonomous driving, which could not have been performed automatically before. There are even applications where deep learning outperformed humans, like object recognition or games. Another field in which this development is showing a huge potential is the medical domain. With the collection of large quantities of patient records and data, and a trend towards personalized treatments, there is a great need for an automatic and reliable processing and analysis of this information. Patient data is not only collected in clinical centres, like hospitals, but it relates also to data coming from general practitioners, healthcare smartphone apps or online websites, just to name a few. This trend resulted in new, massive research efforts during the last years. In Q2/2020, the search engine PubMed returns already over 11.000 results for the search term $'$deep learning$'$, and around 90% of these publications are from the last three years. Hence, a complete overview of the field of $'$medical deep learning$'$ is almost impossible to obtain and getting a full overview of medical sub-fields gets increasingly more difficult. Nevertheless, several review and survey articles about medical deep learning have been presented within the last years. They focused, in general, on specific medical scenarios, like the analysis of medical images containing specific pathologies. With these surveys as foundation, the aim of this contribution is to provide a very first high-level, systematic meta-review of medical deep learning surveys.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge