Henrik Skibbe
Generating visual explanations from deep networks using implicit neural representations
Jan 20, 2025



Abstract:Explaining deep learning models in a way that humans can easily understand is essential for responsible artificial intelligence applications. Attribution methods constitute an important area of explainable deep learning. The attribution problem involves finding parts of the network's input that are the most responsible for the model's output. In this work, we demonstrate that implicit neural representations (INRs) constitute a good framework for generating visual explanations. Firstly, we utilize coordinate-based implicit networks to reformulate and extend the extremal perturbations technique and generate attribution masks. Experimental results confirm the usefulness of our method. For instance, by proper conditioning of the implicit network, we obtain attribution masks that are well-behaved with respect to the imposed area constraints. Secondly, we present an iterative INR-based method that can be used to generate multiple non-overlapping attribution masks for the same image. We depict that a deep learning model may associate the image label with both the appearance of the object of interest as well as with areas and textures usually accompanying the object. Our study demonstrates that implicit networks are well-suited for the generation of attribution masks and can provide interesting insights about the performance of deep learning models.
PatchMorph: A Stochastic Deep Learning Approach for Unsupervised 3D Brain Image Registration with Small Patches
Dec 12, 2023Abstract:We introduce "PatchMorph," an new stochastic deep learning algorithm tailored for unsupervised 3D brain image registration. Unlike other methods, our method uses compact patches of a constant small size to derive solutions that can combine global transformations with local deformations. This approach minimizes the memory footprint of the GPU during training, but also enables us to operate on numerous amounts of randomly overlapping small patches during inference to mitigate image and patch boundary problems. PatchMorph adeptly handles world coordinate transformations between two input images, accommodating variances in attributes such as spacing, array sizes, and orientations. The spatial resolution of patches transitions from coarse to fine, addressing both global and local attributes essential for aligning the images. Each patch offers a unique perspective, together converging towards a comprehensive solution. Experiments on human T1 MRI brain images and marmoset brain images from serial 2-photon tomography affirm PatchMorph's superior performance.
Implicit neural representations for joint decomposition and registration of gene expression images in the marmoset brain
Aug 08, 2023Abstract:We propose a novel image registration method based on implicit neural representations that addresses the challenging problem of registering a pair of brain images with similar anatomical structures, but where one image contains additional features or artifacts that are not present in the other image. To demonstrate its effectiveness, we use 2D microscopy $\textit{in situ}$ hybridization gene expression images of the marmoset brain. Accurately quantifying gene expression requires image registration to a brain template, which is difficult due to the diversity of patterns causing variations in visible anatomical brain structures. Our approach uses implicit networks in combination with an image exclusion loss to jointly perform the registration and decompose the image into a support and residual image. The support image aligns well with the template, while the residual image captures individual image characteristics that diverge from the template. In experiments, our method provided excellent results and outperformed other registration techniques.
Few-shot medical image classification with simple shape and texture text descriptors using vision-language models
Aug 08, 2023



Abstract:In this work, we investigate the usefulness of vision-language models (VLMs) and large language models for binary few-shot classification of medical images. We utilize the GPT-4 model to generate text descriptors that encapsulate the shape and texture characteristics of objects in medical images. Subsequently, these GPT-4 generated descriptors, alongside VLMs pre-trained on natural images, are employed to classify chest X-rays and breast ultrasound images. Our results indicate that few-shot classification of medical images using VLMs and GPT-4 generated descriptors is a viable approach. However, accurate classification requires to exclude certain descriptors from the calculations of the classification scores. Moreover, we assess the ability of VLMs to evaluate shape features in breast mass ultrasound images. We further investigate the degree of variability among the sets of text descriptors produced by GPT-4. Our work provides several important insights about the application of VLMs for medical image analysis.
Improving Segmentation of Objects with Varying Sizes in Biomedical Images using Instance-wise and Center-of-Instance Segmentation Loss Function
Apr 13, 2023



Abstract:In this paper, we propose a novel two-component loss for biomedical image segmentation tasks called the Instance-wise and Center-of-Instance (ICI) loss, a loss function that addresses the instance imbalance problem commonly encountered when using pixel-wise loss functions such as the Dice loss. The Instance-wise component improves the detection of small instances or ``blobs" in image datasets with both large and small instances. The Center-of-Instance component improves the overall detection accuracy. We compared the ICI loss with two existing losses, the Dice loss and the blob loss, in the task of stroke lesion segmentation using the ATLAS R2.0 challenge dataset from MICCAI 2022. Compared to the other losses, the ICI loss provided a better balanced segmentation, and significantly outperformed the Dice loss with an improvement of $1.7-3.7\%$ and the blob loss by $0.6-5.0\%$ in terms of the Dice similarity coefficient on both validation and test set, suggesting that the ICI loss is a potential solution to the instance imbalance problem.
An automated pipeline to create an atlas of in situ hybridization gene expression data in the adult marmoset brain
Mar 13, 2023



Abstract:We present the first automated pipeline to create an atlas of in situ hybridization gene expression in the adult marmoset brain in the same stereotaxic space. The pipeline consists of segmentation of gene expression from microscopy images and registration of images to a standard space. Automation of this pipeline is necessary to analyze the large volume of data in the genome-wide whole-brain dataset, and to process images that have varying intensity profiles and expression patterns with minimal human bias. To reduce the number of labelled images required for training, we develop a semi-supervised segmentation model. We further develop an iterative algorithm to register images to a standard space, enabling comparative analysis between genes and concurrent visualization with other datasets, thereby facilitating a more holistic understanding of primate brain structure and function.
Deep Neural Patchworks: Coping with Large Segmentation Tasks
Jun 07, 2022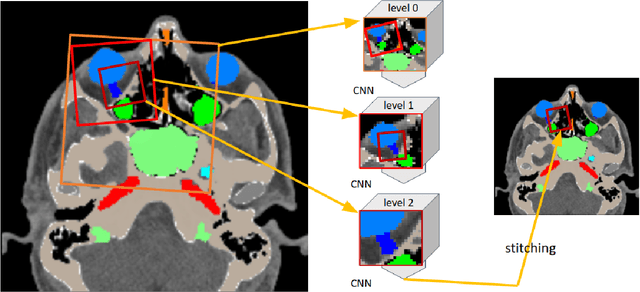
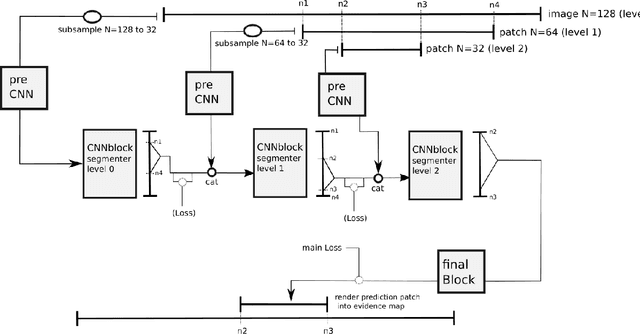
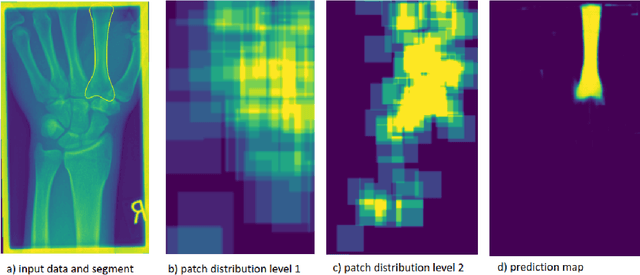
Abstract:Convolutional neural networks are the way to solve arbitrary image segmentation tasks. However, when images are large, memory demands often exceed the available resources, in particular on a common GPU. Especially in biomedical imaging, where 3D images are common, the problems are apparent. A typical approach to solve this limitation is to break the task into smaller subtasks by dividing images into smaller image patches. Another approach, if applicable, is to look at the 2D image sections separately, and to solve the problem in 2D. Often, the loss of global context makes such approaches less effective; important global information might not be present in the current image patch, or the selected 2D image section. Here, we propose Deep Neural Patchworks (DNP), a segmentation framework that is based on hierarchical and nested stacking of patch-based networks that solves the dilemma between global context and memory limitations.
MarmoNet: a pipeline for automated projection mapping of the common marmoset brain from whole-brain serial two-photon tomography
Aug 02, 2019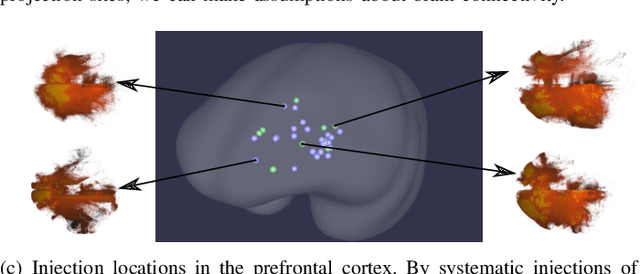
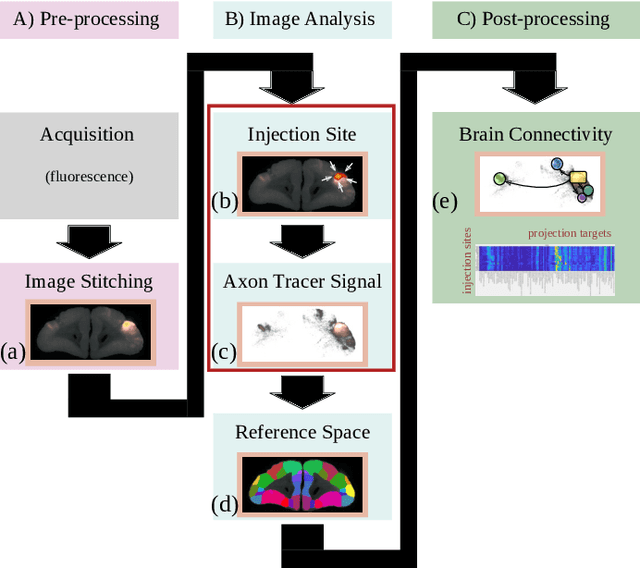
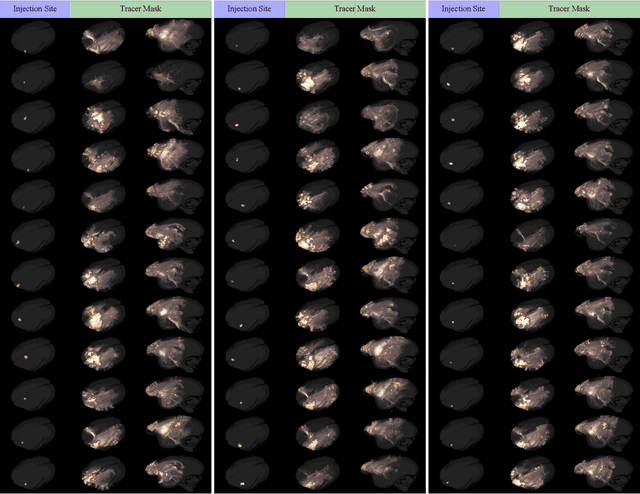
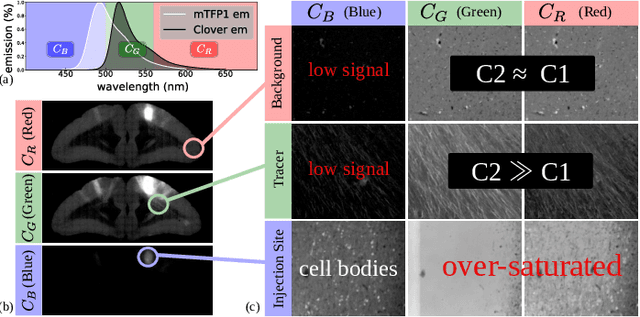
Abstract:Understanding the connectivity in the brain is an important prerequisite for understanding how the brain processes information. In the Brain/MINDS project, a connectivity study on marmoset brains uses two-photon microscopy fluorescence images of axonal projections to collect the neuron connectivity from defined brain regions at the mesoscopic scale. The processing of the images requires the detection and segmentation of the axonal tracer signal. The objective is to detect as much tracer signal as possible while not misclassifying other background structures as the signal. This can be challenging because of imaging noise, a cluttered image background, distortions or varying image contrast cause problems. We are developing MarmoNet, a pipeline that processes and analyzes tracer image data of the common marmoset brain. The pipeline incorporates state-of-the-art machine learning techniques based on artificial convolutional neural networks (CNN) and image registration techniques to extract and map all relevant information in a robust manner. The pipeline processes new images in a fully automated way. This report introduces the current state of the tracer signal analysis part of the pipeline.
HAMLET: Hierarchical Harmonic Filters for Learning Tracts from Diffusion MRI
Jul 03, 2018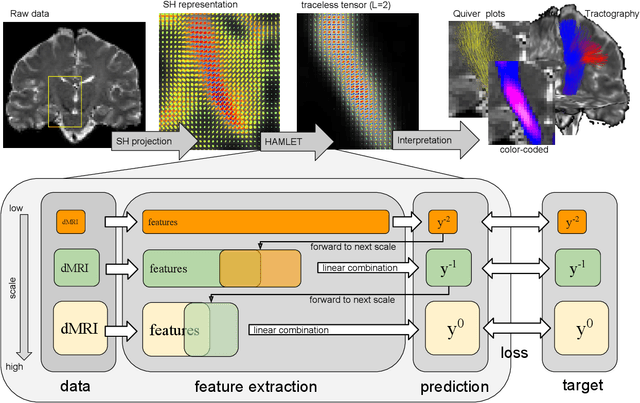
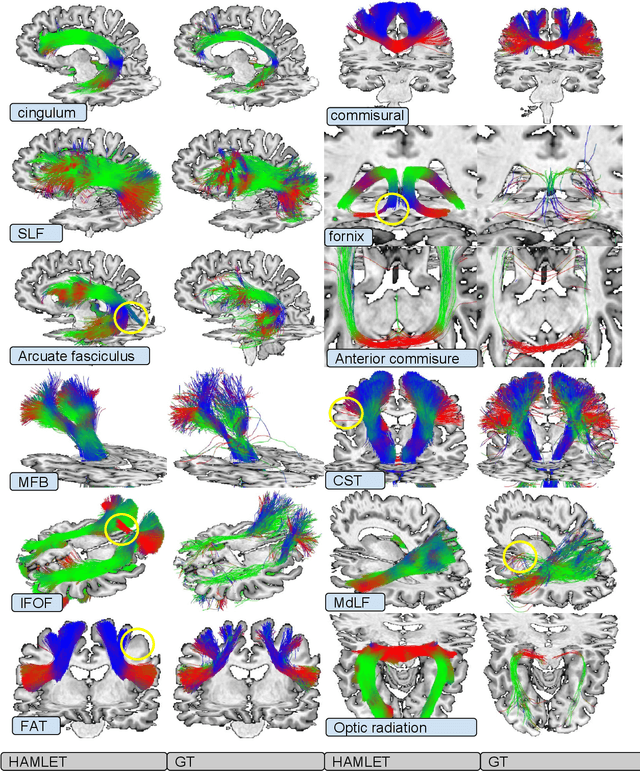
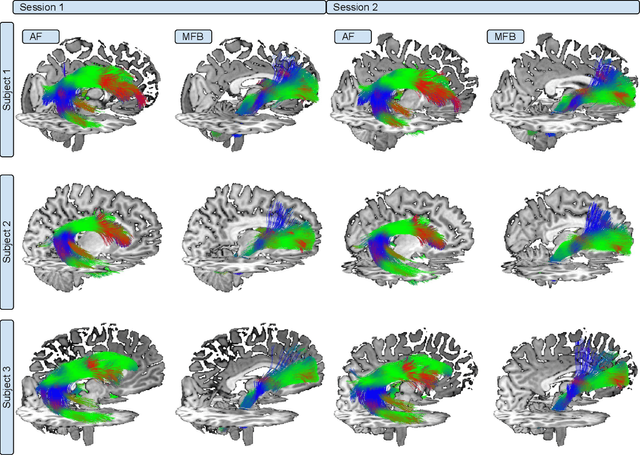
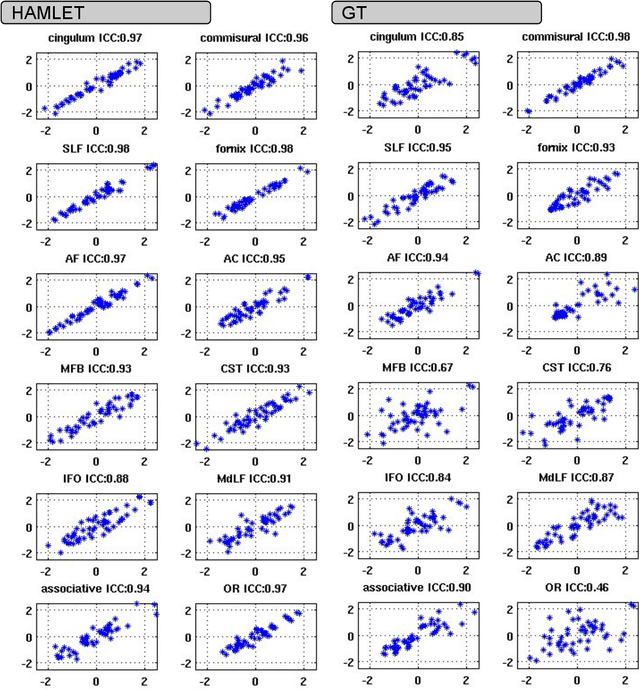
Abstract:In this work we propose HAMLET, a novel tract learning algorithm, which, after training, maps raw diffusion weighted MRI directly onto an image which simultaneously indicates tract direction and tract presence. The automatic learning of fiber tracts based on diffusion MRI data is a rather new idea, which tries to overcome limitations of atlas-based techniques. HAMLET takes a such an approach. Unlike the current trend in machine learning, HAMLET has only a small number of free parameters HAMLET is based on spherical tensor algebra which allows a translation and rotation covariant treatment of the problem. HAMLET is based on a repeated application of convolutions and non-linearities, which all respect the rotation covariance. The intrinsic treatment of such basic image transformations in HAMLET allows the training and generalization of the algorithm without any additional data augmentation. We demonstrate the performance of our approach for twelve prominent bundles, and show that the obtained tract estimates are robust and reliable. It is also shown that the learned models are portable from one sequence to another.
Left-Invariant Diffusion on the Motion Group in terms of the Irreducible Representations of SO
Feb 24, 2012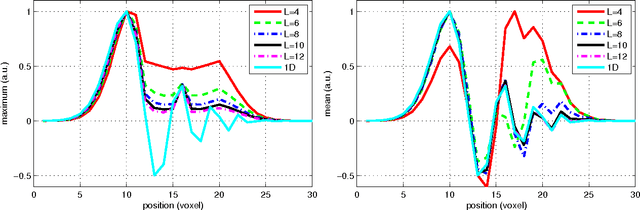
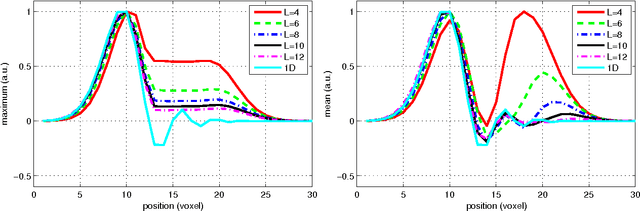
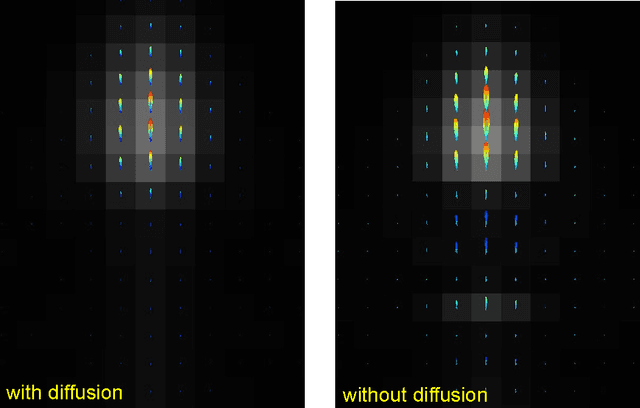
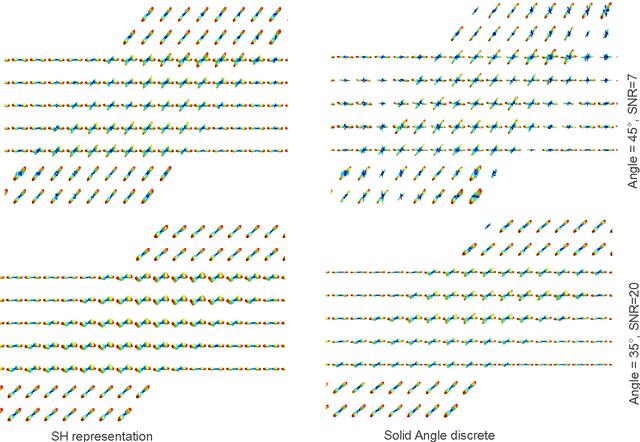
Abstract:In this work we study the formulation of convection/diffusion equations on the 3D motion group SE(3) in terms of the irreducible representations of SO(3). Therefore, the left-invariant vector-fields on SE(3) are expressed as linear operators, that are differential forms in the translation coordinate and algebraic in the rotation. In the context of 3D image processing this approach avoids the explicit discretization of SO(3) or $S_2$, respectively. This is particular important for SO(3), where a direct discretization is infeasible due to the enormous memory consumption. We show two applications of the framework: one in the context of diffusion-weighted magnetic resonance imaging and one in the context of object detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge