Hassan Rivaz
A deep learning approach for patchless estimation of ultrasound quantitative parametric image with uncertainty measurement
Feb 24, 2023Abstract:Quantitative ultrasound (QUS) aims to find properties of scatterers which are related to the tissue microstructure. Among different QUS parameters, scatterer number density has been found to be a reliable biomarker for detecting different abnormalities. The homodyned K-distribution (HK-distribution) is a model for the probability density function of the ultrasound echo amplitude that can model different scattering scenarios but requires a large number of samples to be estimated reliably. Parametric images of HK-distribution parameters can be formed by dividing the envelope data into small overlapping patches and estimating parameters within the patches independently. This approach imposes two limiting constraints, the HK-distribution parameters are assumed to be constant within each patch, and each patch requires enough independent samples. In order to mitigate those problems, we employ a deep learning approach to estimate parametric images of scatterer number density (related to HK-distribution shape parameter) without patching. Furthermore, an uncertainty map of the network's prediction is quantified to provide insight about the confidence of the network about the estimated HK parameter values.
Lateral Strain Imaging using Self-supervised and Physically Inspired Constraints in Unsupervised Regularized Elastography
Dec 16, 2022Abstract:Convolutional Neural Networks (CNN) have shown promising results for displacement estimation in UltraSound Elastography (USE). Many modifications have been proposed to improve the displacement estimation of CNNs for USE in the axial direction. However, the lateral strain, which is essential in several downstream tasks such as the inverse problem of elasticity imaging, remains a challenge. The lateral strain estimation is complicated since the motion and the sampling frequency in this direction are substantially lower than the axial one, and a lack of carrier signal in this direction. In computer vision applications, the axial and the lateral motions are independent. In contrast, the tissue motion pattern in USE is governed by laws of physics which link the axial and lateral displacements. In this paper, inspired by Hooke's law, we first propose Physically Inspired ConsTraint for Unsupervised Regularized Elastography (PICTURE), where we impose a constraint on the Effective Poisson's ratio (EPR) to improve the lateral strain estimation. In the next step, we propose self-supervised PICTURE (sPICTURE) to further enhance the strain image estimation. Extensive experiments on simulation, experimental phantom and in vivo data demonstrate that the proposed methods estimate accurate axial and lateral strain maps.
Infusing known operators in convolutional neural networks for lateral strain imaging in ultrasound elastography
Oct 31, 2022Abstract:Convolutional Neural Networks (CNN) have been employed for displacement estimation in ultrasound elastography (USE). High-quality axial strains (derivative of the axial displacement in the axial direction) can be estimated by the proposed networks. In contrast to axial strain, lateral strain, which is highly required in Poisson's ratio imaging and elasticity reconstruction, has a poor quality. The main causes include low sampling frequency, limited motion, and lack of phase information in the lateral direction. Recently, physically inspired constraint in unsupervised regularized elastography (PICTURE) has been proposed. This method took into account the range of the feasible lateral strain defined by the rules of physics of motion and employed a regularization strategy to improve the lateral strains. Despite the substantial improvement, the regularization was only applied during the training; hence it did not guarantee during the test that the lateral strain is within the feasible range. Furthermore, only the feasible range was employed, other constraints such as incompressibility were not investigated. In this paper, we address these two issues and propose kPICTURE in which two iterative algorithms were infused into the network architecture in the form of known operators to ensure the lateral strain is within the feasible range and impose incompressibility during the test phase.
Homodyned K-distribution: parameter estimation and uncertainty quantification using Bayesian neural networks
Oct 31, 2022Abstract:Quantitative ultrasound (QUS) allows estimating the intrinsic tissue properties. Speckle statistics are the QUS parameters that describe the first order statistics of ultrasound (US) envelope data. The parameters of Homodyned K-distribution (HK-distribution) are the speckle statistics that can model the envelope data in diverse scattering conditions. However, they require a large amount of data to be estimated reliably. Consequently, finding out the intrinsic uncertainty of the estimated parameters can help us to have a better understanding of the estimated parameters. In this paper, we propose a Bayesian Neural Network (BNN) to estimate the parameters of HK-distribution and quantify the uncertainty of the estimator.
Transformer-Based Microbubble Localization
Sep 23, 2022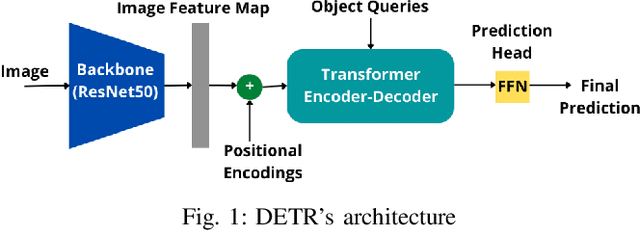
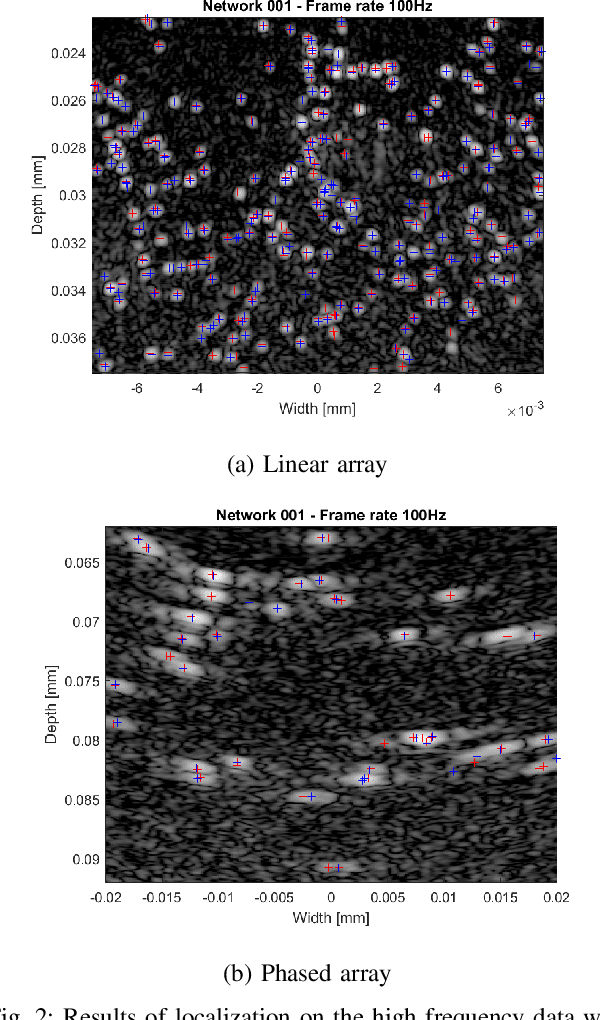
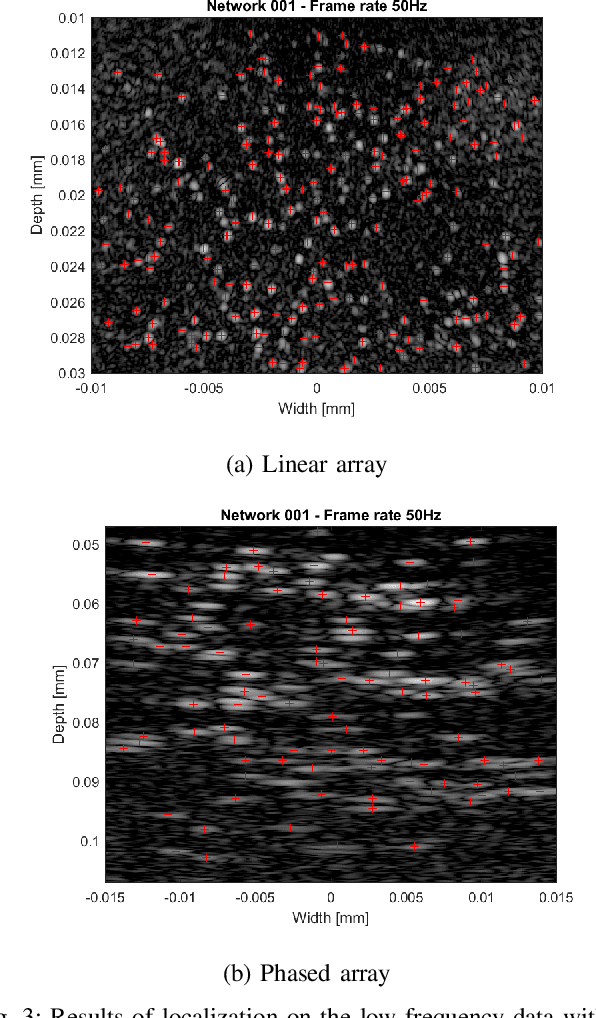
Abstract:Ultrasound Localization Microscopy (ULM) is an emerging technique that employs the localization of echogenic microbubbles (MBs) to finely sample and image the microcirculation beyond the diffraction limit of ultrasound imaging. Conventional MB localization methods are mainly based on considering a specific Point Spread Function (PSF) for MBs, which leads to loss of information caused by overlapping MBs, non-stationary PSFs, and harmonic MB echoes. Therefore, it is imperative to devise methods that can accurately localize MBs while being resilient to MB nonlinearities and variations of MB concentrations that distort MB PSFs. This paper proposes a transformer-based MB localization approach to address this issue. We adopted DEtection TRansformer (DETR) arXiv:2005.12872 , which is an end-to-end object recognition method that detects a unique bounding box for each of the detected objects using set-based Hungarian loss and bipartite matching. To the authors' knowledge, this is the first time transformers have been used for MB localization. To appraise the proposed strategy, the pre-trained DETR network's performance has been tested for detecting MBs using transfer learning principles. We have fine-tuned the network on a subset of randomly selected frames of the dataset provided by the IEEE IUS Ultra-SR challenge organizers and then tested on the rest using cross-validation. For the simulation dataset, the paper supports the deployment of transformer-based solutions for MB localization at high accuracy.
Analytic Optimization-Based Microbubble Tracking in Ultrasound Super-Resolution Microscopy
Sep 21, 2022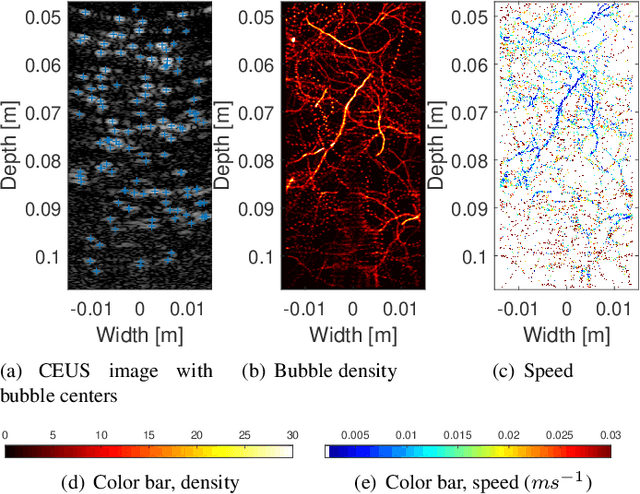
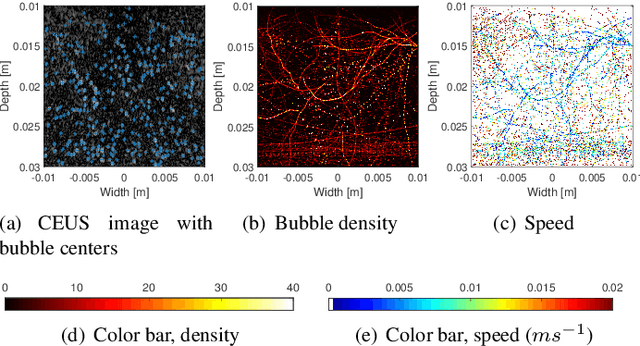
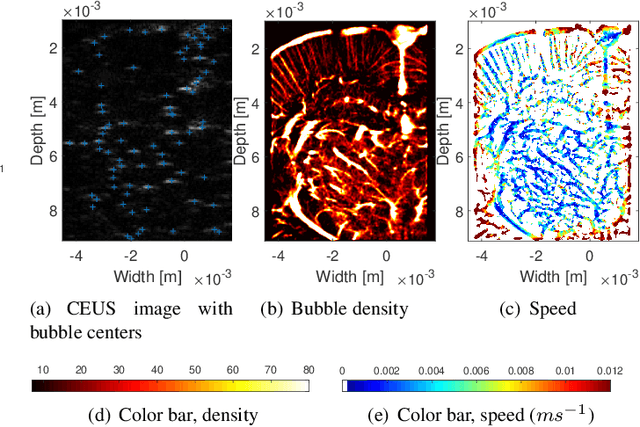
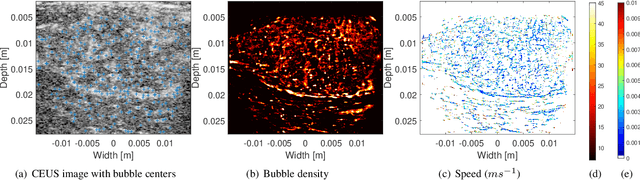
Abstract:Ultrasound localization microscopy (ULM) refers to a promising medical imaging modality that systematically leverages the advantages of contrast-enhanced ultrasound (CEUS) to surpass the diffraction barrier and delineate the microvascular map. Localization and tracking of microbubbles (MBs), two significant steps of ULM, facilitate generating the vascular map and the velocity distribution, respectively. Herein, we propose a novel MB tracking technique considering temporal pairing as a bubble-set registration problem. Iterative registration is performed between the bubble sets in two consecutive time instants by analytically optimizing a cost function that takes position and point-spread function (PSF) similarities as well as physically plausible levels of bubbles' movement into account. Furthermore, we infer MBs' parity in a fuzzy manner instead of binary. The proposed technique performs well in validation experiments with two synthetic and two in vivo datasets provided by the Ultrasound Localisation and TRacking Algorithms for Super Resolution (ULTRA-SR) Challenge.
DiffeoRaptor: Diffeomorphic Inter-modal Image Registration using RaPTOR
Sep 12, 2022
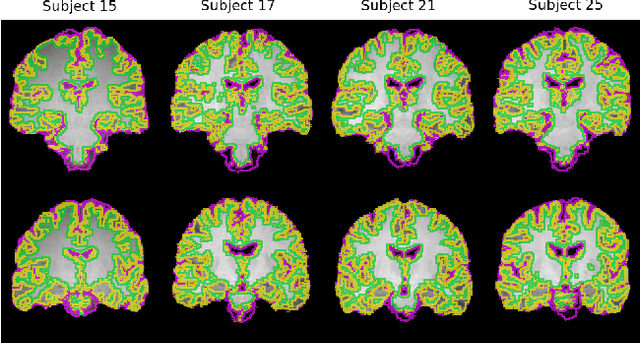
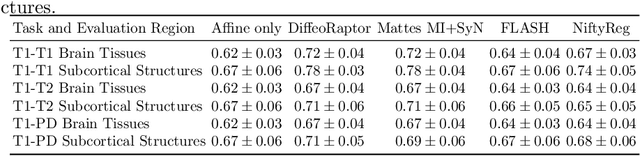
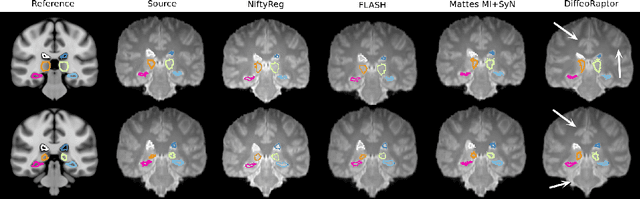
Abstract:Purpose: Diffeomorphic image registration is essential in many medical imaging applications. Several registration algorithms of such type have been proposed, but primarily for intra-contrast alignment. Currently, efficient inter-modal/contrast diffeomorphic registration, which is vital in numerous applications, remains a challenging task. Methods: We proposed a novel inter-modal/contrast registration algorithm that leverages Robust PaTch-based cOrrelation Ratio (RaPTOR) metric to allow inter-modal/contrast image alignment and bandlimited geodesic shooting demonstrated in Fourier Approximated Lie Algebras (FLASH) algorithm for fast diffeomorphic registration. Results: The proposed algorithm, named DiffeoRaptor, was validated with three public databases for the tasks of brain and abdominal image registration while comparing the results against three state-of-the-art techniques, including FLASH, NiftyReg, and Symmetric image normalization (SyN). Conclusions: Our results demonstrated that DiffeoRaptor offered comparable or better registration performance in terms of registration accuracy. Moreover, DiffeoRaptor produces smoother deformations than SyN in inter-modal and contrast registration. The code for DiffeoRaptor is publicly available at https://github.com/nimamasoumi/DiffeoRaptor.
Physics-Inspired Regularized Pulse-Echo Quantitative Ultrasound: Efficient Optimization with ADMM
Aug 13, 2022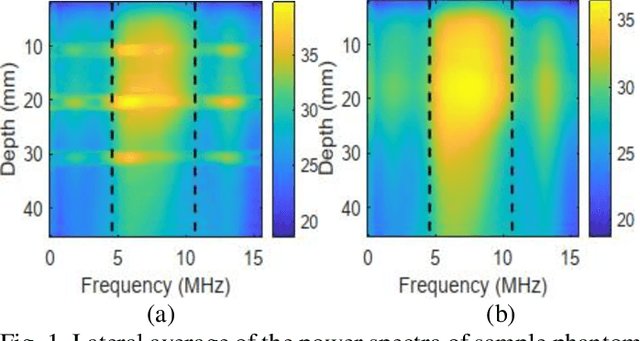
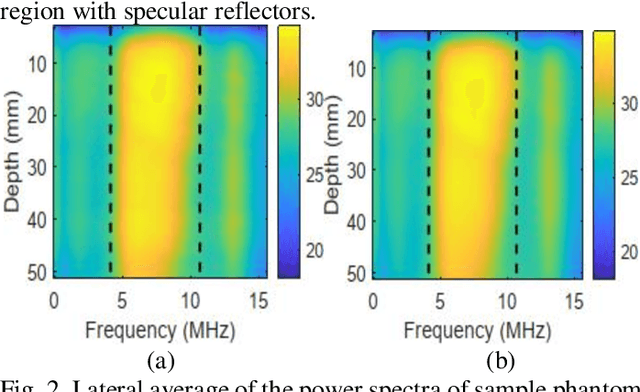
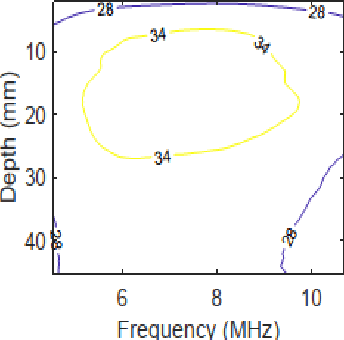
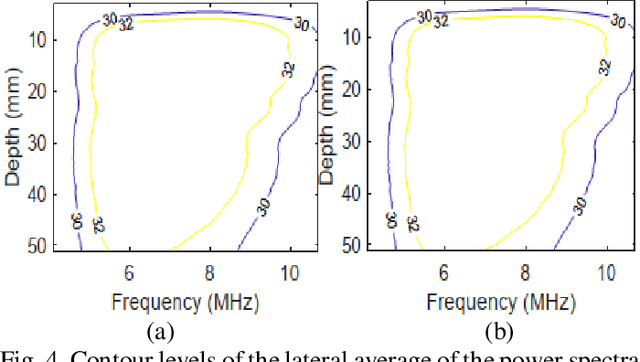
Abstract:Pulse-echo Quantitative ultrasound (PEQUS), which estimates the quantitative properties of tissue microstructure, entails estimating the average attenuation and the backscatter coefficient (BSC). Growing recent research has focused on the regularized estimation of these parameters. Herein, we make two contributions to this field: First, we consider the physics of the average attenuation and backscattering to devise regularization terms accordingly. More specifically, since the average attenuation gradually alters in different parts of the tissue while BSC can vary markedly from tissue to tissue, we apply L2 and L1 norms for the average attenuation and the BSC, respectively. Second, we multiply different frequencies and depths of the power spectra with different weights according to their noise levels. Our rationale is that the high-frequency contents of the power spectra at deep regions have a low signal-to-noise ratio. We exploit the alternating direction method of multipliers (ADMM) for optimizing the cost function. Qualitative and quantitative evaluation of bias and variance exhibit that our proposed algorithm substantially improves the estimations of the average attenuation and the BSC.
RESECT-SEG: Open access annotations of intra-operative brain tumor ultrasound images
Jul 13, 2022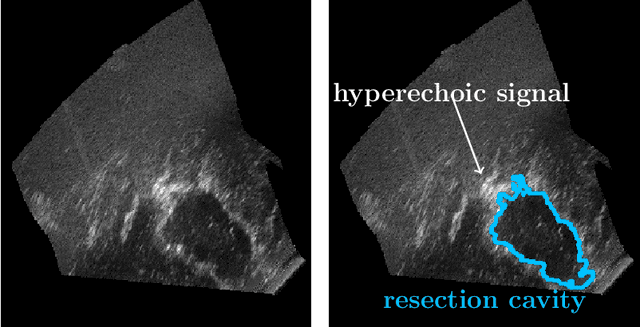
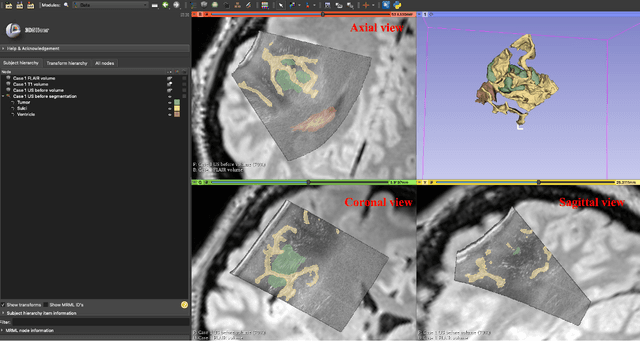
Abstract:Purpose: Registration and segmentation of magnetic resonance (MR) and ultrasound (US) images play an essential role in surgical planning and resection of brain tumors. However, validating these techniques is challenging due to the scarcity of publicly accessible sources with high-quality ground truth information. To this end, we propose a unique annotation dataset of tumor tissues and resection cavities from the previously published RESECT dataset (Xiao et al. 2017) to encourage a more rigorous assessments of image processing techniques. Acquisition and validation methods: The RESECT database consists of MR and intraoperative US (iUS) images of 23 patients who underwent resection surgeries. The proposed dataset contains tumor tissues and resection cavity annotations of the iUS images. The quality of annotations were validated by two highly experienced neurosurgeons through several assessment criteria. Data format and availability: Annotations of tumor tissues and resection cavities are provided in 3D NIFTI formats. Both sets of annotations are accessible online in the \url{https://osf.io/6y4db}. Discussion and potential applications: The proposed database includes tumor tissue and resection cavity annotations from real-world clinical ultrasound brain images to evaluate segmentation and registration methods. These labels could also be used to train deep learning approaches. Eventually, this dataset should further improve the quality of image guidance in neurosurgery.
Inverse Problem of Ultrasound Beamforming with Denoising-Based Regularized Solutions
Jun 16, 2022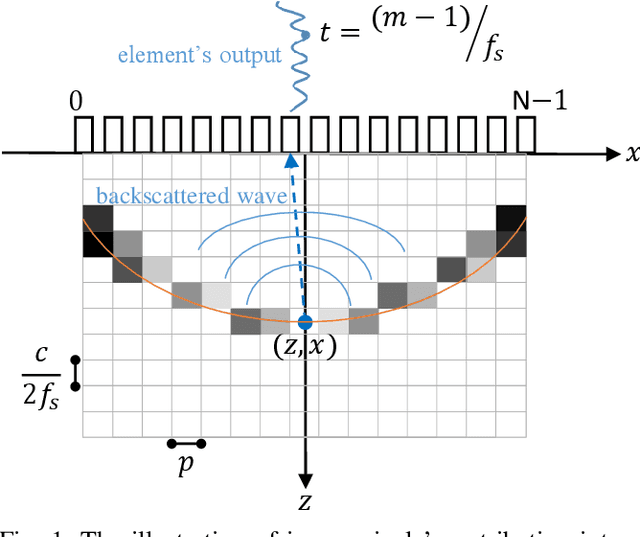
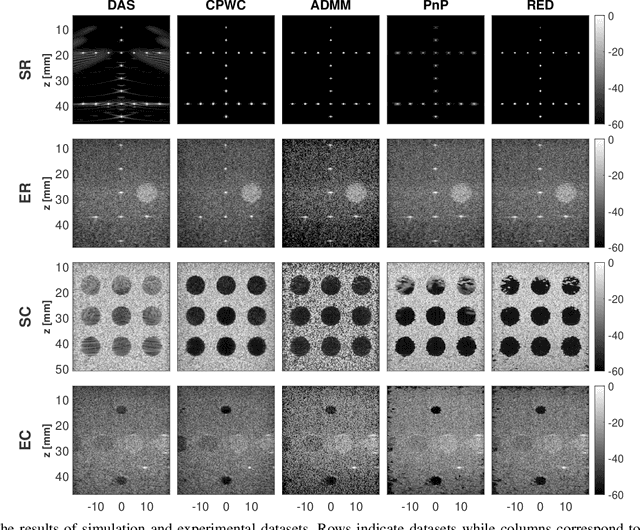
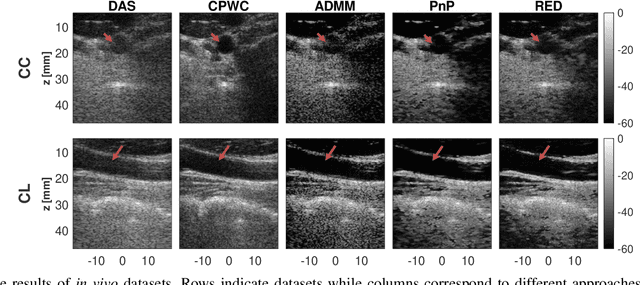
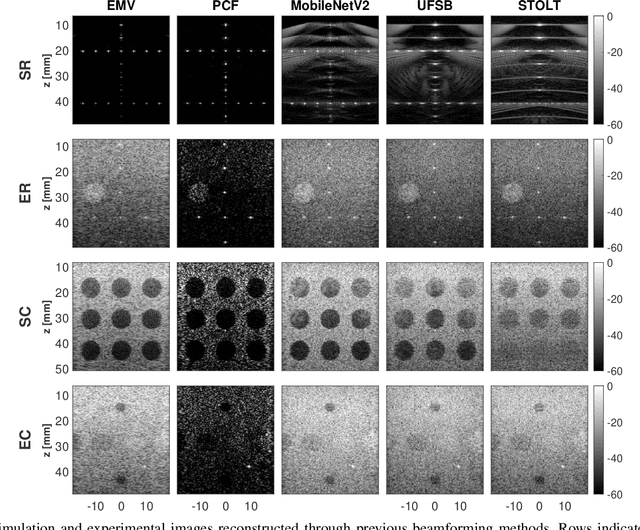
Abstract:During the past few years, inverse problem formulations of ultrasound beamforming have attracted a growing interest. They usually pose beamforming as a minimization problem of a fidelity term resulting from the measurement model plus a regularization term that enforces a certain class on the resulting image. Herein, we take advantages of alternating direction method of multipliers to propose a flexible framework in which each term is optimized separately. Furthermore, the proposed beamforming formulation is extended to replace the regularization term by a denoising algorithm, based on the recent approaches called plug-and-play (PnP) and regularization by denoising (RED). Such regularizations are shown in this work to better preserve speckle texture, an important feature in ultrasound imaging, than sparsity-based approaches previously proposed in the literature. The efficiency of proposed methods is evaluated on simulations, real phantoms, and \textit{in vivo} data available from a plane-wave imaging challenge in medical ultrasound. Furthermore, a comprehensive comparison with existing ultrasound beamforming methods is also provided. These results show that the RED algorithm gives the best image quality in terms of contrast index while preserving the speckle statistics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge