Haoyu Pan
Universal Battery Degradation Forecasting Driven by Foundation Model Across Diverse Chemistries and Conditions
Dec 30, 2025Abstract:Accurate forecasting of battery capacity fade is essential for the safety, reliability, and long-term efficiency of energy storage systems. However, the strong heterogeneity across cell chemistries, form factors, and operating conditions makes it difficult to build a single model that generalizes beyond its training domain. This work proposes a unified capacity forecasting framework that maintains robust performance across diverse chemistries and usage scenarios. We curate 20 public aging datasets into a large-scale corpus covering 1,704 cells and 3,961,195 charge-discharge cycle segments, spanning temperatures from $-5\,^{\circ}\mathrm{C}$ to $45\,^{\circ}\mathrm{C}$, multiple C-rates, and application-oriented profiles such as fast charging and partial cycling. On this corpus, we adopt a Time-Series Foundation Model (TSFM) backbone and apply parameter-efficient Low-Rank Adaptation (LoRA) together with physics-guided contrastive representation learning to capture shared degradation patterns. Experiments on both seen and deliberately held-out unseen datasets show that a single unified model achieves competitive or superior accuracy compared with strong per-dataset baselines, while retaining stable performance on chemistries, capacity scales, and operating conditions excluded from training. These results demonstrate the potential of TSFM-based architectures as a scalable and transferable solution for capacity degradation forecasting in real battery management systems.
Breast Ultrasound Tumor Generation via Mask Generator and Text-Guided Network:A Clinically Controllable Framework with Downstream Evaluation
Jul 10, 2025Abstract:The development of robust deep learning models for breast ultrasound (BUS) image analysis is significantly constrained by the scarcity of expert-annotated data. To address this limitation, we propose a clinically controllable generative framework for synthesizing BUS images. This framework integrates clinical descriptions with structural masks to generate tumors, enabling fine-grained control over tumor characteristics such as morphology, echogencity, and shape. Furthermore, we design a semantic-curvature mask generator, which synthesizes structurally diverse tumor masks guided by clinical priors. During inference, synthetic tumor masks serve as input to the generative framework, producing highly personalized synthetic BUS images with tumors that reflect real-world morphological diversity. Quantitative evaluations on six public BUS datasets demonstrate the significant clinical utility of our synthetic images, showing their effectiveness in enhancing downstream breast cancer diagnosis tasks. Furthermore, visual Turing tests conducted by experienced sonographers confirm the realism of the generated images, indicating the framework's potential to support broader clinical applications.
Enhancing Spatiotemporal Prediction Model using Modular Design and Beyond
Oct 04, 2022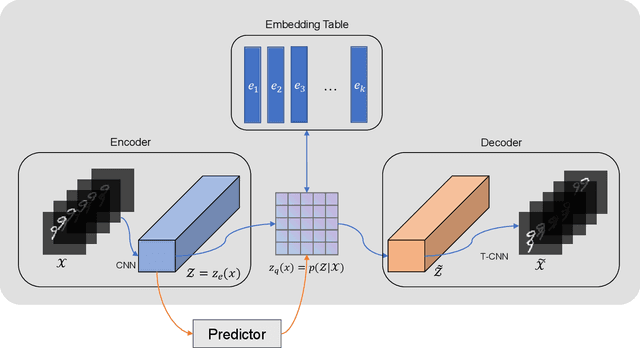
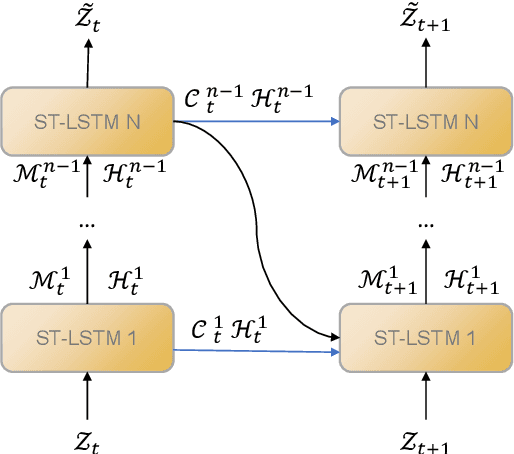
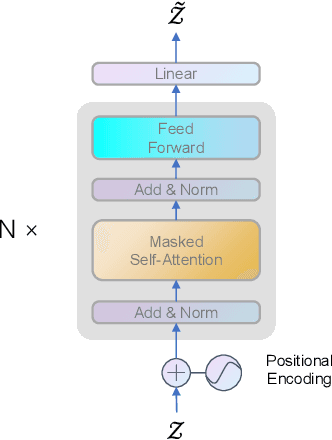

Abstract:Predictive learning uses a known state to generate a future state over a period of time. It is a challenging task to predict spatiotemporal sequence because the spatiotemporal sequence varies both in time and space. The mainstream method is to model spatial and temporal structures at the same time using RNN-based or transformer-based architecture, and then generates future data by using learned experience in the way of auto-regressive. The method of learning spatial and temporal features simultaneously brings a lot of parameters to the model, which makes the model difficult to be convergent. In this paper, a modular design is proposed, which decomposes spatiotemporal sequence model into two modules: a spatial encoder-decoder and a predictor. These two modules can extract spatial features and predict future data respectively. The spatial encoder-decoder maps the data into a latent embedding space and generates data from the latent space while the predictor forecasts future embedding from past. By applying the design to the current research and performing experiments on KTH-Action and MovingMNIST datasets, we both improve computational performance and obtain state-of-the-art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge