Hans J. Grabe
DenseNet and Support Vector Machine classifications of major depressive disorder using vertex-wise cortical features
Nov 18, 2023



Abstract:Major depressive disorder (MDD) is a complex psychiatric disorder that affects the lives of hundreds of millions of individuals around the globe. Even today, researchers debate if morphological alterations in the brain are linked to MDD, likely due to the heterogeneity of this disorder. The application of deep learning tools to neuroimaging data, capable of capturing complex non-linear patterns, has the potential to provide diagnostic and predictive biomarkers for MDD. However, previous attempts to demarcate MDD patients and healthy controls (HC) based on segmented cortical features via linear machine learning approaches have reported low accuracies. In this study, we used globally representative data from the ENIGMA-MDD working group containing an extensive sample of people with MDD (N=2,772) and HC (N=4,240), which allows a comprehensive analysis with generalizable results. Based on the hypothesis that integration of vertex-wise cortical features can improve classification performance, we evaluated the classification of a DenseNet and a Support Vector Machine (SVM), with the expectation that the former would outperform the latter. As we analyzed a multi-site sample, we additionally applied the ComBat harmonization tool to remove potential nuisance effects of site. We found that both classifiers exhibited close to chance performance (balanced accuracy DenseNet: 51%; SVM: 53%), when estimated on unseen sites. Slightly higher classification performance (balanced accuracy DenseNet: 58%; SVM: 55%) was found when the cross-validation folds contained subjects from all sites, indicating site effect. In conclusion, the integration of vertex-wise morphometric features and the use of the non-linear classifier did not lead to the differentiability between MDD and HC. Our results support the notion that MDD classification on this combination of features and classifiers is unfeasible.
Medical Image Harmonization Using Deep Learning Based Canonical Mapping: Toward Robust and Generalizable Learning in Imaging
Oct 11, 2020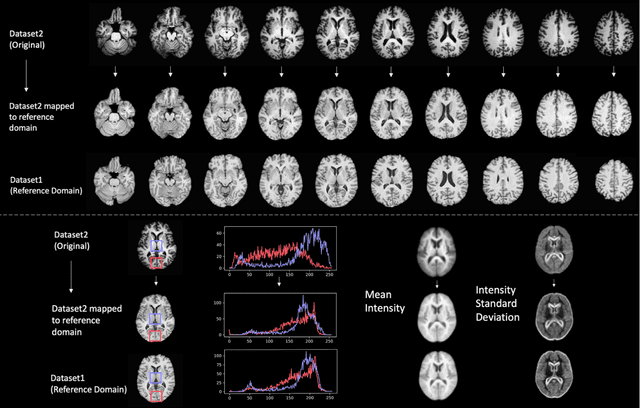
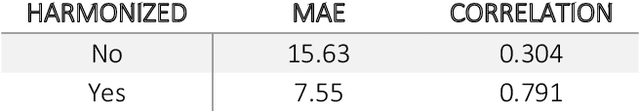
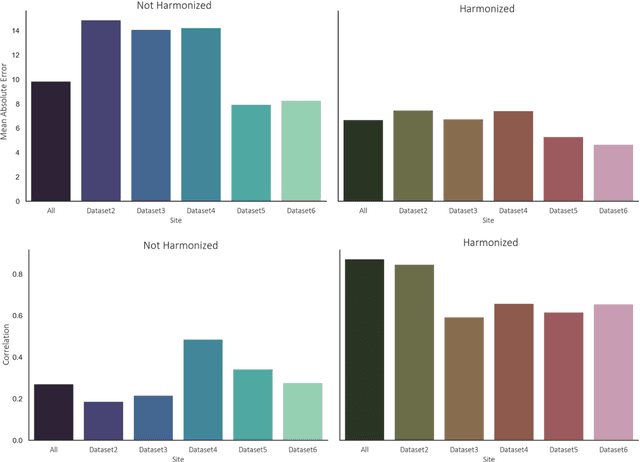
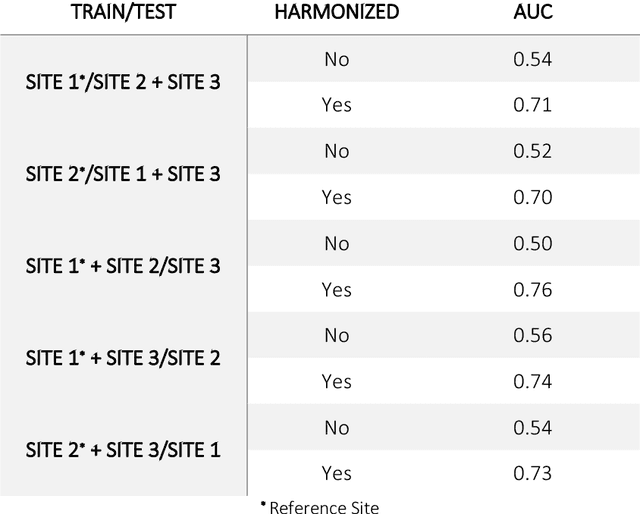
Abstract:Conventional and deep learning-based methods have shown great potential in the medical imaging domain, as means for deriving diagnostic, prognostic, and predictive biomarkers, and by contributing to precision medicine. However, these methods have yet to see widespread clinical adoption, in part due to limited generalization performance across various imaging devices, acquisition protocols, and patient populations. In this work, we propose a new paradigm in which data from a diverse range of acquisition conditions are "harmonized" to a common reference domain, where accurate model learning and prediction can take place. By learning an unsupervised image to image canonical mapping from diverse datasets to a reference domain using generative deep learning models, we aim to reduce confounding data variation while preserving semantic information, thereby rendering the learning task easier in the reference domain. We test this approach on two example problems, namely MRI-based brain age prediction and classification of schizophrenia, leveraging pooled cohorts of neuroimaging MRI data spanning 9 sites and 9701 subjects. Our results indicate a substantial improvement in these tasks in out-of-sample data, even when training is restricted to a single site.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge