Gunjan Balde
Evaluation of LLMs in Medical Text Summarization: The Role of Vocabulary Adaptation in High OOV Settings
May 27, 2025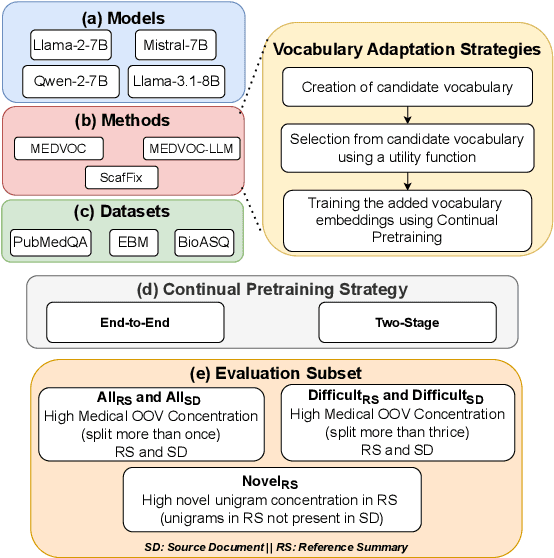
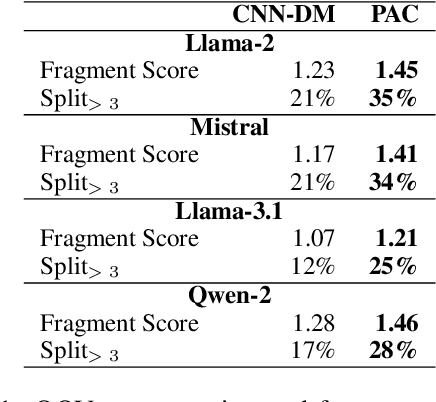
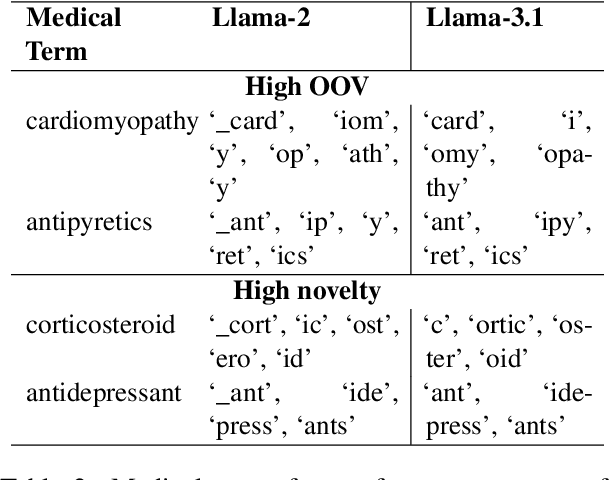
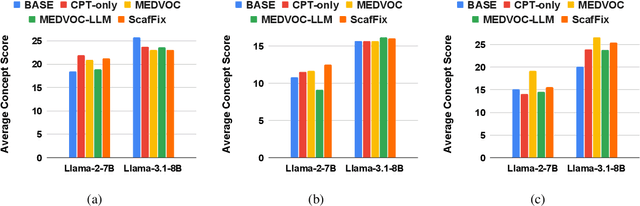
Abstract:Large Language Models (LLMs) recently achieved great success in medical text summarization by simply using in-context learning. However, these recent efforts do not perform fine-grained evaluations under difficult settings where LLMs might fail. They typically report performance scores over the entire dataset. Through our benchmarking study, we show that LLMs show a significant performance drop for data points with high concentration of out-of-vocabulary (OOV) words or with high novelty. Vocabulary adaptation is an intuitive solution to this vocabulary mismatch issue where the LLM vocabulary gets updated with certain expert domain (here, medical) words or subwords. An interesting finding from our study is that Llama-3.1, even with a vocabulary size of around 128K tokens, still faces over-fragmentation issue with medical words. To that end, we show vocabulary adaptation helps improve the LLM summarization performance even in difficult settings. Through extensive experimentation of multiple vocabulary adaptation strategies, two continual pretraining strategies, and three benchmark medical summarization datasets, we gain valuable insights into the role of vocabulary adaptation strategies for customizing LLMs to the medical domain. We also performed a human evaluation study with medical experts where they found that vocabulary adaptation results in more relevant and faithful summaries. Our codebase is made publicly available at https://github.com/gb-kgp/LLM-MedicalSummarization-Benchmark.
Adaptive BPE Tokenization for Enhanced Vocabulary Adaptation in Finetuning Pretrained Language Models
Oct 04, 2024



Abstract:In this work, we show a fundamental limitation in vocabulary adaptation approaches that use Byte-Pair Encoding (BPE) tokenization scheme for fine-tuning pretrained language models (PLMs) to expert domains. Current approaches trivially append the target domain-specific vocabulary at the end of the PLM vocabulary. This approach leads to a lower priority score and causes sub-optimal tokenization in BPE that iteratively uses merge rules to tokenize a given text. To mitigate this issue, we propose AdaptBPE where the BPE tokenization initialization phase is modified to first perform the longest string matching on the added (target) vocabulary before tokenizing at the character level. We perform an extensive evaluation of AdaptBPE versus the standard BPE over various classification and summarization tasks; AdaptBPE improves by 3.57% (in terms of accuracy) and 1.87% (in terms of Rouge-L), respectively. AdaptBPE for MEDVOC works particularly well when reference summaries have high OOV concentration or are longer in length. We also conduct a human evaluation, revealing that AdaptBPE generates more relevant and more faithful summaries as compared to MEDVOC. We make our codebase publicly available at https://github.com/gb-kgp/adaptbpe.
MEDVOC: Vocabulary Adaptation for Fine-tuning Pre-trained Language Models on Medical Text Summarization
May 07, 2024



Abstract:This work presents a dynamic vocabulary adaptation strategy, MEDVOC, for fine-tuning pre-trained language models (PLMs) like BertSumAbs, BART, and PEGASUS for improved medical text summarization. In contrast to existing domain adaptation approaches in summarization, MEDVOC treats vocabulary as an optimizable parameter and optimizes the PLM vocabulary based on fragment score conditioned only on the downstream task's reference summaries. Unlike previous works on vocabulary adaptation (limited only to classification tasks), optimizing vocabulary based on summarization tasks requires an extremely costly intermediate fine-tuning step on large summarization datasets. To that end, our novel fragment score-based hyperparameter search very significantly reduces this fine-tuning time -- from 450 days to less than 2 days on average. Furthermore, while previous works on vocabulary adaptation are often primarily tied to single PLMs, MEDVOC is designed to be deployable across multiple PLMs (with varying model vocabulary sizes, pre-training objectives, and model sizes) -- bridging the limited vocabulary overlap between the biomedical literature domain and PLMs. MEDVOC outperforms baselines by 15.74% in terms of Rouge-L in zero-shot setting and shows gains of 17.29% in high Out-Of-Vocabulary (OOV) concentrations. Our human evaluation shows MEDVOC generates more faithful medical summaries (88% compared to 59% in baselines). We make the codebase publicly available at https://github.com/gb-kgp/MEDVOC.
Knowledge-Aware Neural Networks for Medical Forum Question Classification
Sep 27, 2021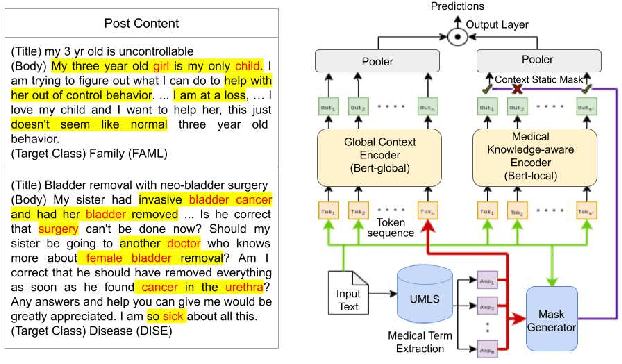
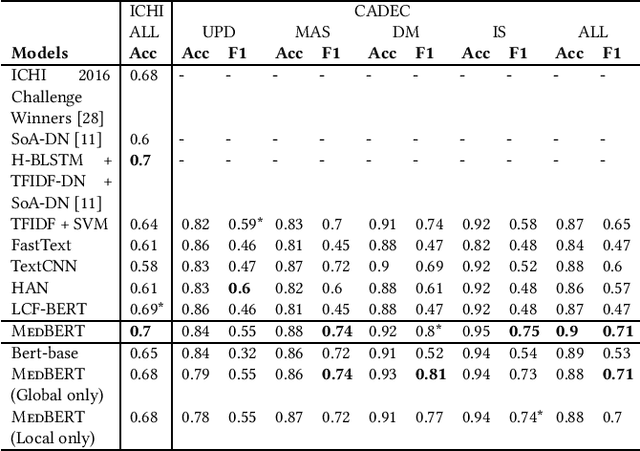
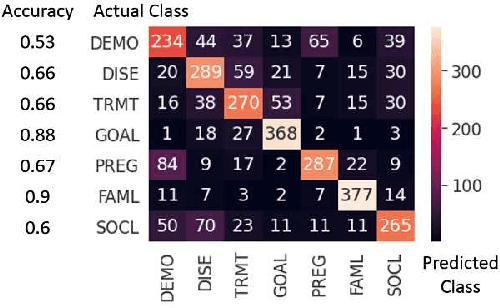
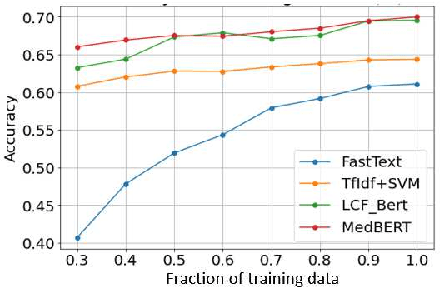
Abstract:Online medical forums have become a predominant platform for answering health-related information needs of consumers. However, with a significant rise in the number of queries and the limited availability of experts, it is necessary to automatically classify medical queries based on a consumer's intention, so that these questions may be directed to the right set of medical experts. Here, we develop a novel medical knowledge-aware BERT-based model (MedBERT) that explicitly gives more weightage to medical concept-bearing words, and utilize domain-specific side information obtained from a popular medical knowledge base. We also contribute a multi-label dataset for the Medical Forum Question Classification (MFQC) task. MedBERT achieves state-of-the-art performance on two benchmark datasets and performs very well in low resource settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge