Georgia Tourassi
Adiabatic Quantum Support Vector Machines
Jan 23, 2024Abstract:Adiabatic quantum computers can solve difficult optimization problems (e.g., the quadratic unconstrained binary optimization problem), and they seem well suited to train machine learning models. In this paper, we describe an adiabatic quantum approach for training support vector machines. We show that the time complexity of our quantum approach is an order of magnitude better than the classical approach. Next, we compare the test accuracy of our quantum approach against a classical approach that uses the Scikit-learn library in Python across five benchmark datasets (Iris, Wisconsin Breast Cancer (WBC), Wine, Digits, and Lambeq). We show that our quantum approach obtains accuracies on par with the classical approach. Finally, we perform a scalability study in which we compute the total training times of the quantum approach and the classical approach with increasing number of features and number of data points in the training dataset. Our scalability results show that the quantum approach obtains a 3.5--4.5 times speedup over the classical approach on datasets with many (millions of) features.
Topological Interpretability for Deep-Learning
May 15, 2023Abstract:With the increasing adoption of AI-based systems across everyday life, the need to understand their decision-making mechanisms is correspondingly accelerating. The level at which we can trust the statistical inferences made from AI-based decision systems is an increasing concern, especially in high-risk systems such as criminal justice or medical diagnosis, where incorrect inferences may have tragic consequences. Despite their successes in providing solutions to problems involving real-world data, deep learning (DL) models cannot quantify the certainty of their predictions. And are frequently quite confident, even when their solutions are incorrect. This work presents a method to infer prominent features in two DL classification models trained on clinical and non-clinical text by employing techniques from topological and geometric data analysis. We create a graph of a model's prediction space and cluster the inputs into the graph's vertices by the similarity of features and prediction statistics. We then extract subgraphs demonstrating high-predictive accuracy for a given label. These subgraphs contain a wealth of information about features that the DL model has recognized as relevant to its decisions. We infer these features for a given label using a distance metric between probability measures, and demonstrate the stability of our method compared to the LIME interpretability method. This work demonstrates that we may gain insights into the decision mechanism of a DL model, which allows us to ascertain if the model is making its decisions based on information germane to the problem or identifies extraneous patterns within the data.
Integration of Domain Knowledge using Medical Knowledge Graph Deep Learning for Cancer Phenotyping
Jan 05, 2021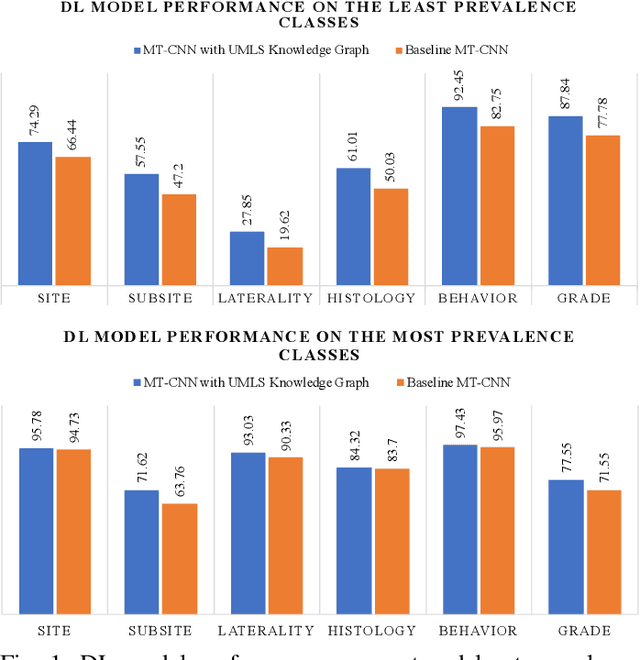
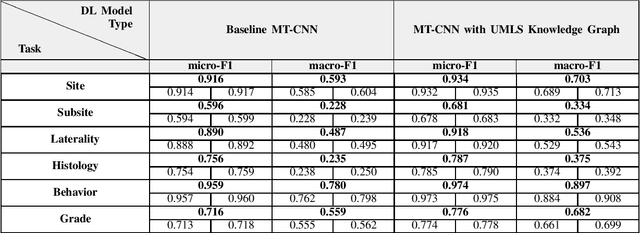
Abstract:A key component of deep learning (DL) for natural language processing (NLP) is word embeddings. Word embeddings that effectively capture the meaning and context of the word that they represent can significantly improve the performance of downstream DL models for various NLP tasks. Many existing word embeddings techniques capture the context of words based on word co-occurrence in documents and text; however, they often cannot capture broader domain-specific relationships between concepts that may be crucial for the NLP task at hand. In this paper, we propose a method to integrate external knowledge from medical terminology ontologies into the context captured by word embeddings. Specifically, we use a medical knowledge graph, such as the unified medical language system (UMLS), to find connections between clinical terms in cancer pathology reports. This approach aims to minimize the distance between connected clinical concepts. We evaluate the proposed approach using a Multitask Convolutional Neural Network (MT-CNN) to extract six cancer characteristics -- site, subsite, laterality, behavior, histology, and grade -- from a dataset of ~900K cancer pathology reports. The results show that the MT-CNN model which uses our domain informed embeddings outperforms the same MT-CNN using standard word2vec embeddings across all tasks, with an improvement in the overall micro- and macro-F1 scores by 4.97\%and 22.5\%, respectively.
Why I'm not Answering: Understanding Determinants of Classification of an Abstaining Classifier for Cancer Pathology Reports
Sep 24, 2020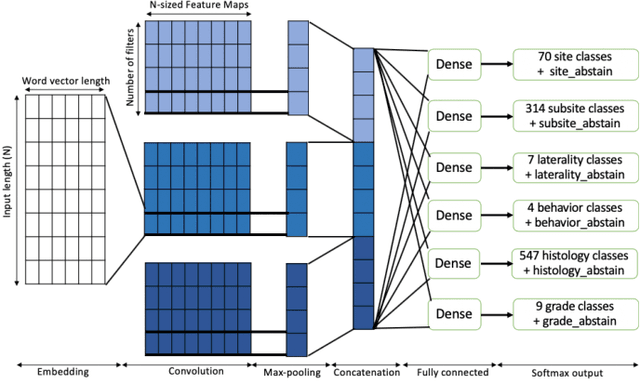
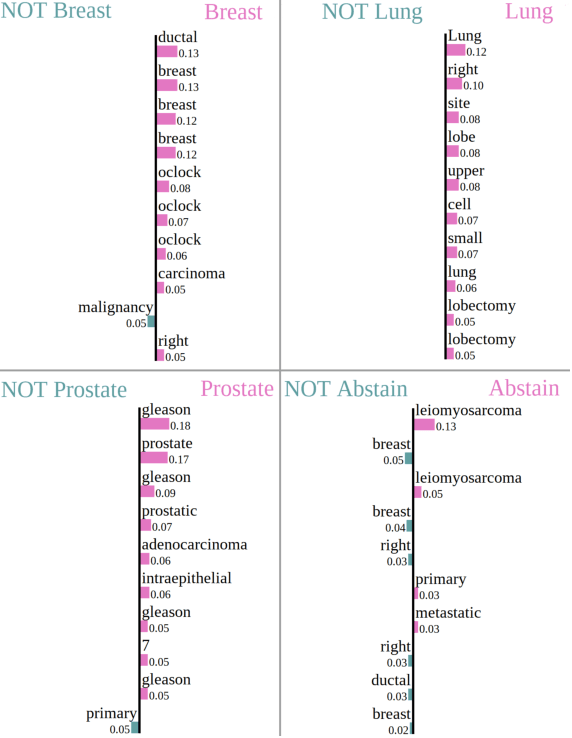
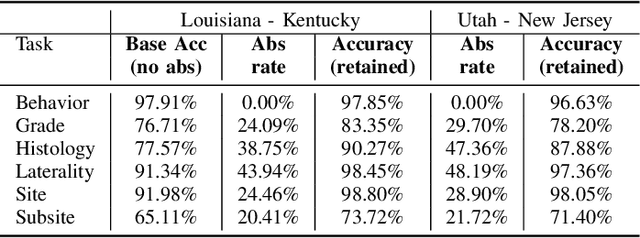
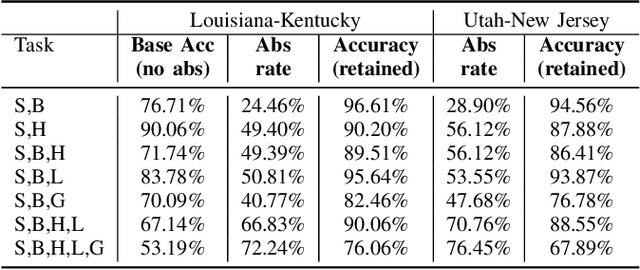
Abstract:Safe deployment of deep learning systems in critical real world applications requires models to make few mistakes, and only under predictable circumstances. Development of such a model is not yet possible, in general. In this work, we address this problem with an abstaining classifier tuned to have $>$95% accuracy, and identify the determinants of abstention with LIME (the Local Interpretable Model-agnostic Explanations method). Essentially, we are training our model to learn the attributes of pathology reports that are likely to lead to incorrect classifications, albeit at the cost of reduced sensitivity. We demonstrate our method in a multitask setting to classify cancer pathology reports from the NCI SEER cancer registries on six tasks of greatest importance. For these tasks, we reduce the classification error rate by factors of 2-5 by abstaining on 25-45% of the reports. For the specific case of cancer site, we are able to identify metastasis and reports involving lymph nodes as responsible for many of the classification mistakes, and that the extent and types of mistakes vary systematically with cancer site (eg. breast, lung, and prostate). When combining across three of the tasks, our model classifies 50% of the reports with an accuracy greater than 95% for three of the six tasks and greater than 85% for all six tasks on the retained samples. By using this information, we expect to define work flows that incorporate machine learning only in the areas where it is sufficiently robust and accurate, saving human attention to areas where it is required.
Precision Medicine as an Accelerator for Next Generation Cognitive Supercomputing
Apr 29, 2018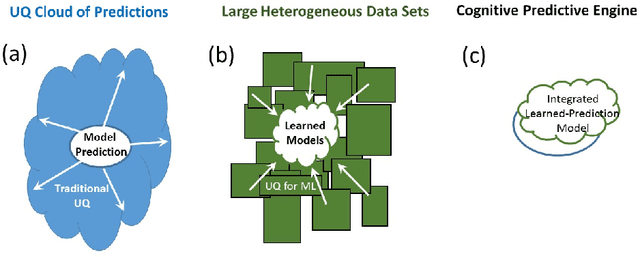
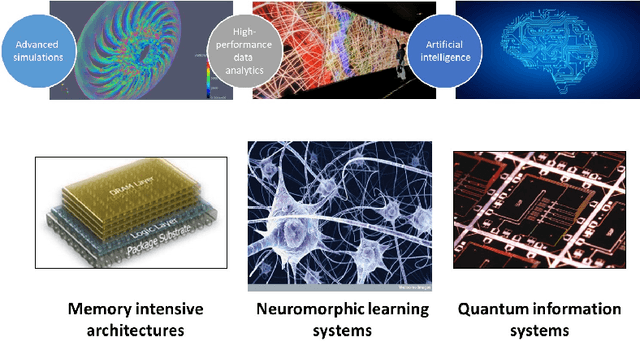
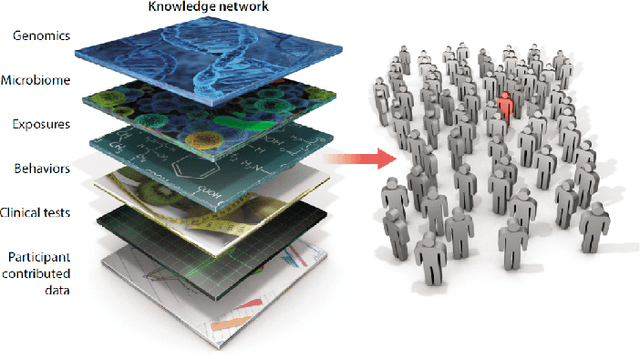
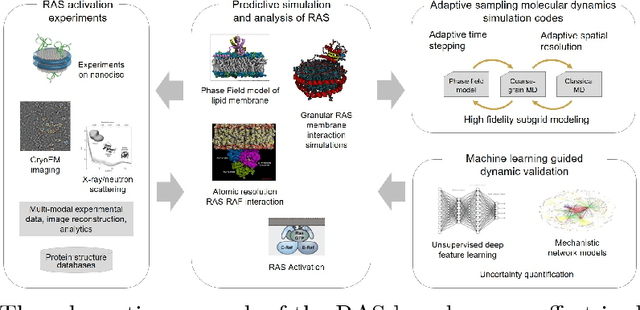
Abstract:In the past several years, we have taken advantage of a number of opportunities to advance the intersection of next generation high-performance computing AI and big data technologies through partnerships in precision medicine. Today we are in the throes of piecing together what is likely the most unique convergence of medical data and computer technologies. But more deeply, we observe that the traditional paradigm of computer simulation and prediction needs fundamental revision. This is the time for a number of reasons. We will review what the drivers are, why now, how this has been approached over the past several years, and where we are heading.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge