Fangfang Xia
Division of Data Science and Learning, Argonne National Laboratory, Argonne, IL, USA, University of Chicago Consortium for Advanced Science and Engineering, University of Chicago, Chicago, IL, USA
Ensemble Transfer Learning for the Prediction of Anti-Cancer Drug Response
May 13, 2020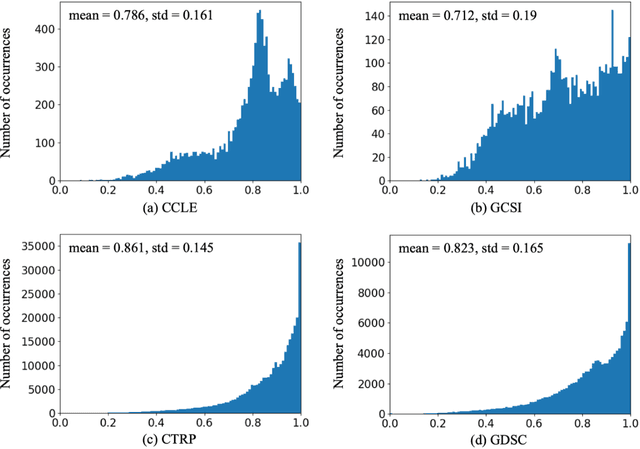
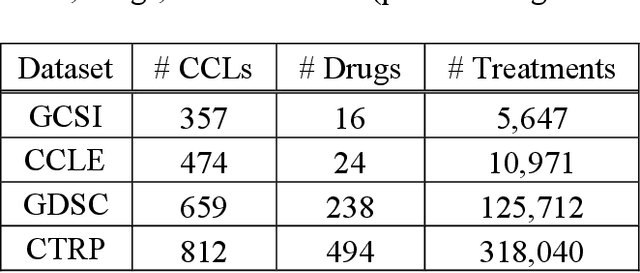
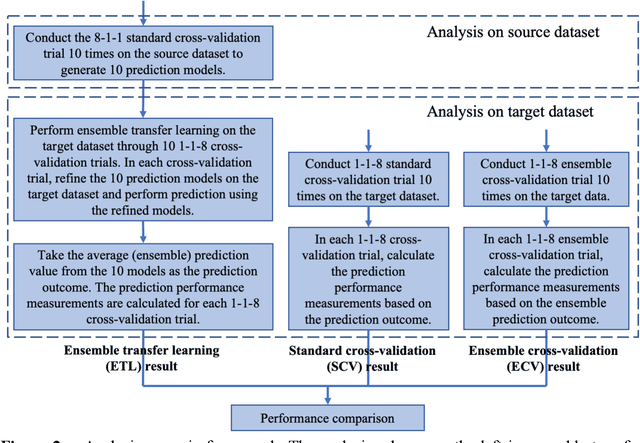
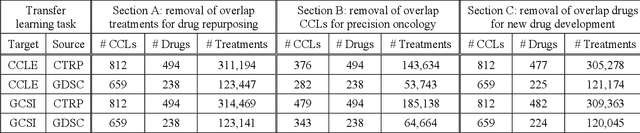
Abstract:Transfer learning has been shown to be effective in many applications in which training data for the target problem are limited but data for a related (source) problem are abundant. In this paper, we apply transfer learning to the prediction of anti-cancer drug response. Previous transfer learning studies for drug response prediction focused on building models that predict the response of tumor cells to a specific drug treatment. We target the more challenging task of building general prediction models that can make predictions for both new tumor cells and new drugs. We apply the classic transfer learning framework that trains a prediction model on the source dataset and refines it on the target dataset, and extends the framework through ensemble. The ensemble transfer learning pipeline is implemented using LightGBM and two deep neural network (DNN) models with different architectures. Uniquely, we investigate its power for three application settings including drug repurposing, precision oncology, and new drug development, through different data partition schemes in cross-validation. We test the proposed ensemble transfer learning on benchmark in vitro drug screening datasets, taking one dataset as the source domain and another dataset as the target domain. The analysis results demonstrate the benefit of applying ensemble transfer learning for predicting anti-cancer drug response in all three applications with both LightGBM and DNN models. Compared between the different prediction models, a DNN model with two subnetworks for the inputs of tumor features and drug features separately outperforms LightGBM and the other DNN model that concatenates tumor features and drug features for input in the drug repurposing and precision oncology applications. In the more challenging application of new drug development, LightGBM performs better than the other two DNN models.
Deep Medical Image Analysis with Representation Learning and Neuromorphic Computing
May 11, 2020
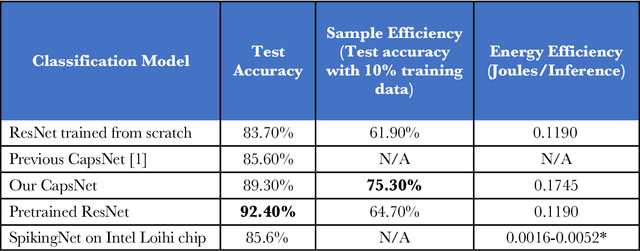


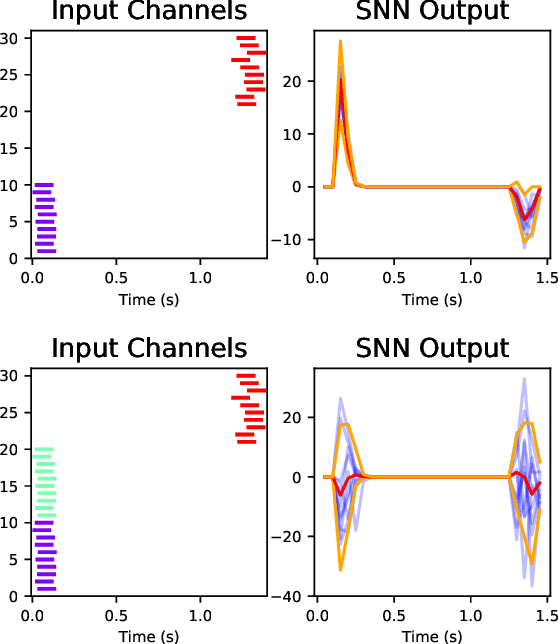
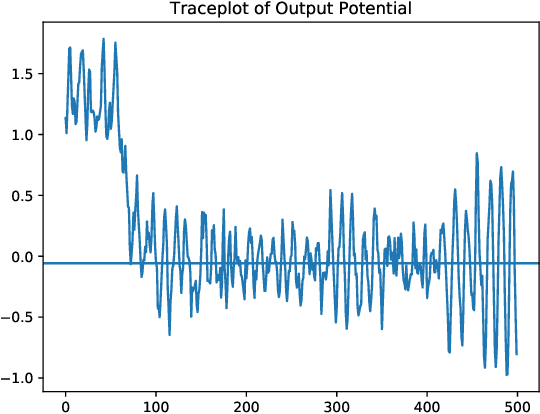
Abstract:We explore three representative lines of research and demonstrate the utility of our methods on a classification benchmark of brain cancer MRI data. First, we present a capsule network that explicitly learns a representation robust to rotation and affine transformation. This model requires less training data and outperforms both the original convolutional baseline and a previous capsule network implementation. Second, we leverage the latest domain adaptation techniques to achieve a new state-of-the-art accuracy. Our experiments show that non-medical images can be used to improve model performance. Finally, we design a spiking neural network trained on the Intel Loihi neuromorphic chip (Fig. 1 shows an inference snapshot). This model consumes much lower power while achieving reasonable accuracy given model reduction. We posit that more research in this direction combining hardware and learning advancements will power future medical imaging (on-device AI, few-shot prediction, adaptive scanning).
Towards On-Chip Bayesian Neuromorphic Learning
May 05, 2020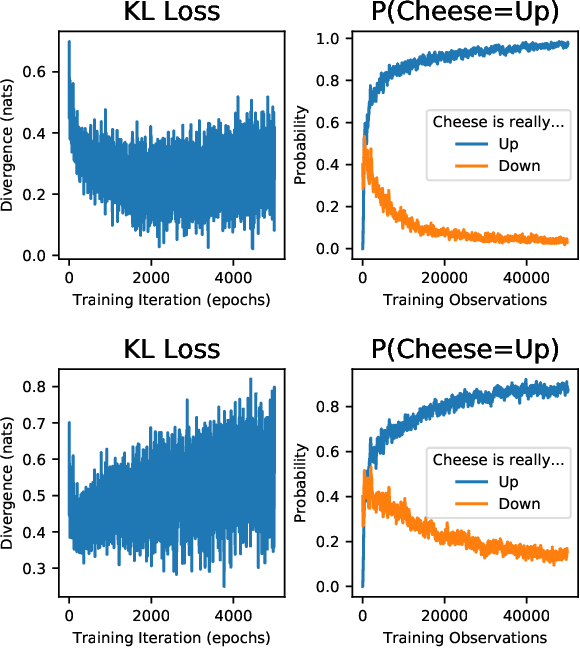
Abstract:If edge devices are to be deployed to critical applications where their decisions could have serious financial, political, or public-health consequences, they will need a way to signal when they are not sure how to react to their environment. For instance, a lost delivery drone could make its way back to a distribution center or contact the client if it is confused about how exactly to make its delivery, rather than taking the action which is "most likely" correct. This issue is compounded for health care or military applications. However, the brain-realistic temporal credit assignment problem neuromorphic computing algorithms have to solve is difficult. The double role weights play in backpropagation-based-learning, dictating how the network reacts to both input and feedback, needs to be decoupled. e-prop 1 is a promising learning algorithm that tackles this with Broadcast Alignment (a technique where network weights are replaced with random weights during feedback) and accumulated local information. We investigate under what conditions the Bayesian loss term can be expressed in a similar fashion, proposing an algorithm that can be computed with only local information as well and which is thus no more difficult to implement on hardware. This algorithm is exhibited on a store-recall problem, which suggests that it can learn good uncertainty on decisions to be made over time.
A Systematic Approach to Featurization for Cancer Drug Sensitivity Predictions with Deep Learning
May 04, 2020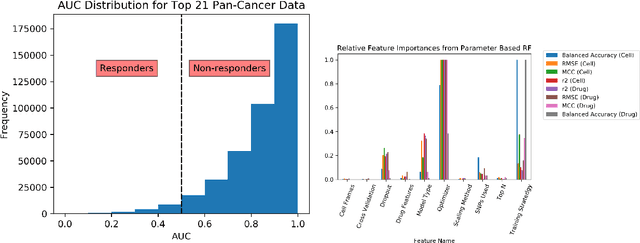
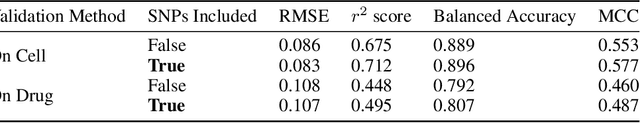
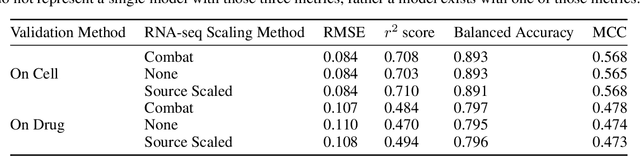
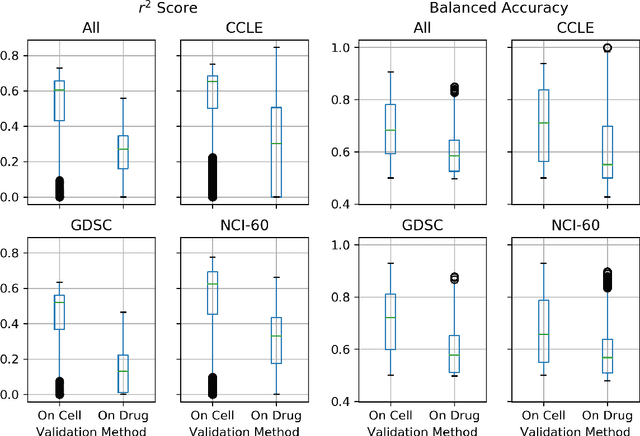
Abstract:By combining various cancer cell line (CCL) drug screening panels, the size of the data has grown significantly to begin understanding how advances in deep learning can advance drug response predictions. In this paper we train >35,000 neural network models, sweeping over common featurization techniques. We found the RNA-seq to be highly redundant and informative even with subsets larger than 128 features. We found the inclusion of single nucleotide polymorphisms (SNPs) coded as count matrices improved model performance significantly, and no substantial difference in model performance with respect to molecular featurization between the common open source MOrdred descriptors and Dragon7 descriptors. Alongside this analysis, we outline data integration between CCL screening datasets and present evidence that new metrics and imbalanced data techniques, as well as advances in data standardization, need to be developed.
Scalable Reinforcement-Learning-Based Neural Architecture Search for Cancer Deep Learning Research
Sep 01, 2019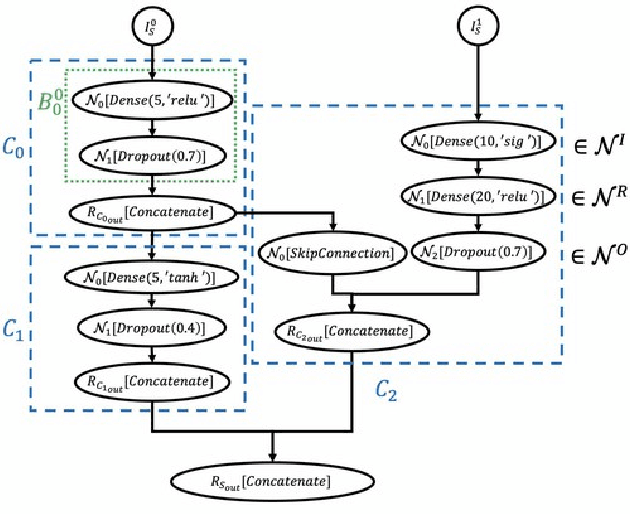
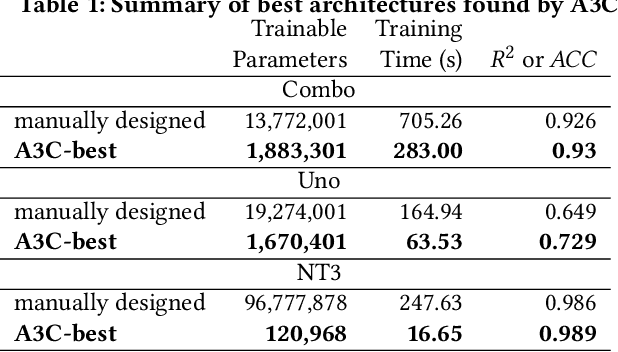
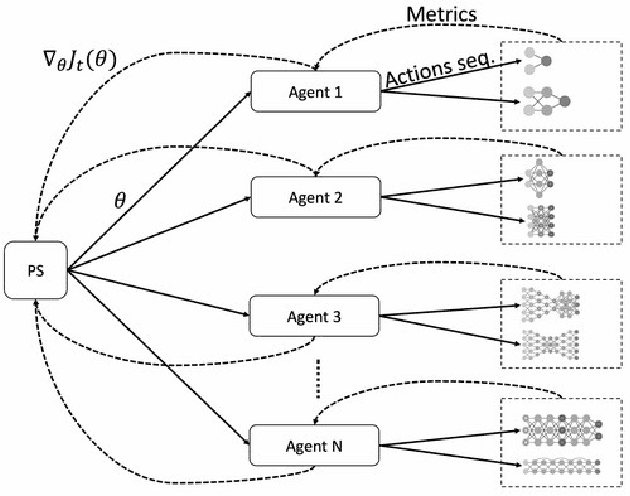
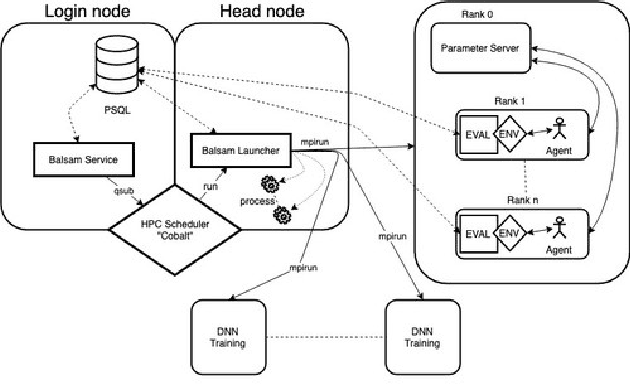
Abstract:Cancer is a complex disease, the understanding and treatment of which are being aided through increases in the volume of collected data and in the scale of deployed computing power. Consequently, there is a growing need for the development of data-driven and, in particular, deep learning methods for various tasks such as cancer diagnosis, detection, prognosis, and prediction. Despite recent successes, however, designing high-performing deep learning models for nonimage and nontext cancer data is a time-consuming, trial-and-error, manual task that requires both cancer domain and deep learning expertise. To that end, we develop a reinforcement-learning-based neural architecture search to automate deep-learning-based predictive model development for a class of representative cancer data. We develop custom building blocks that allow domain experts to incorporate the cancer-data-specific characteristics. We show that our approach discovers deep neural network architectures that have significantly fewer trainable parameters, shorter training time, and accuracy similar to or higher than those of manually designed architectures. We study and demonstrate the scalability of our approach on up to 1,024 Intel Knights Landing nodes of the Theta supercomputer at the Argonne Leadership Computing Facility.
Neuromorphic Acceleration for Approximate Bayesian Inference on Neural Networks via Permanent Dropout
Apr 29, 2019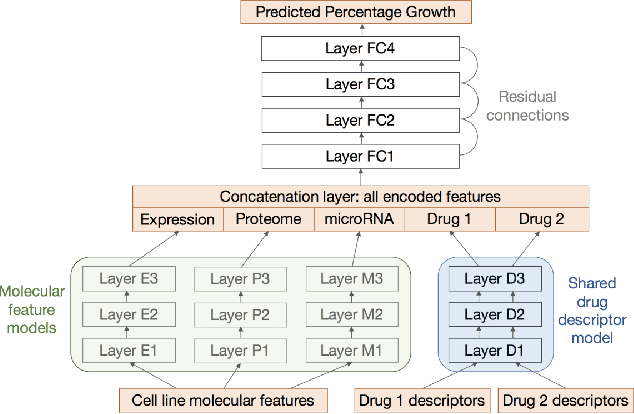
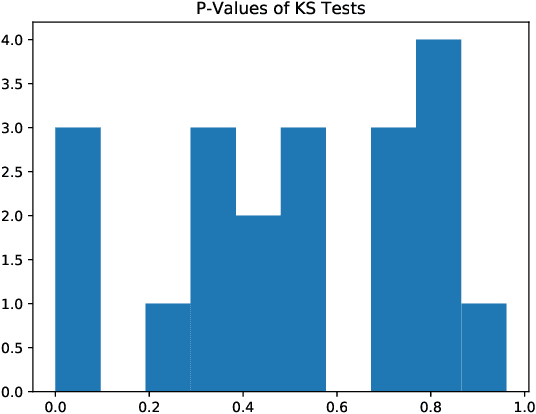
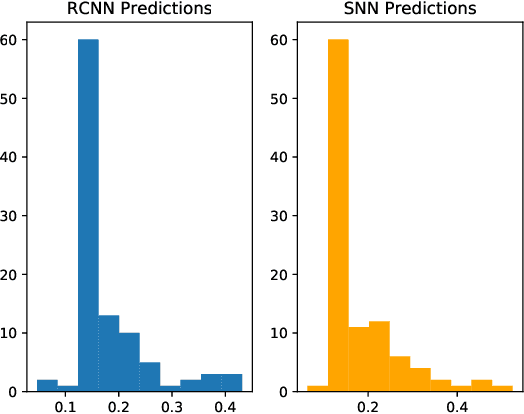
Abstract:As neural networks have begun performing increasingly critical tasks for society, ranging from driving cars to identifying candidates for drug development, the value of their ability to perform uncertainty quantification (UQ) in their predictions has risen commensurately. Permanent dropout, a popular method for neural network UQ, involves injecting stochasticity into the inference phase of the model and creating many predictions for each of the test data. This shifts the computational and energy burden of deep neural networks from the training phase to the inference phase. Recent work has demonstrated near-lossless conversion of classical deep neural networks to their spiking counterparts. We use these results to demonstrate the feasibility of conducting the inference phase with permanent dropout on spiking neural networks, mitigating the technique's computational and energy burden, which is essential for its use at scale or on edge platforms. We demonstrate the proposed approach via the Nengo spiking neural simulator on a combination drug therapy dataset for cancer treatment, where UQ is critical. Our results indicate that the spiking approximation gives a predictive distribution practically indistinguishable from that given by the classical network.
Machine Learning for Antimicrobial Resistance
Jul 05, 2016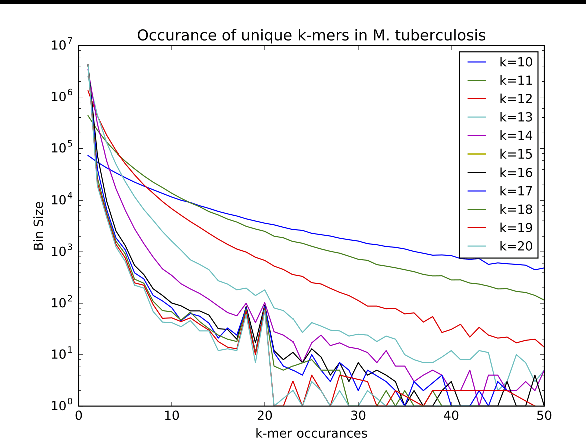
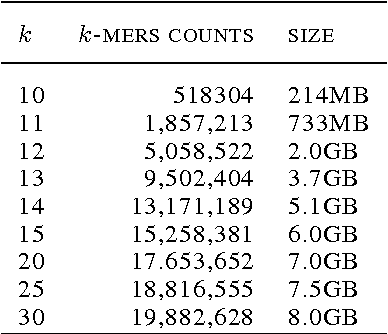
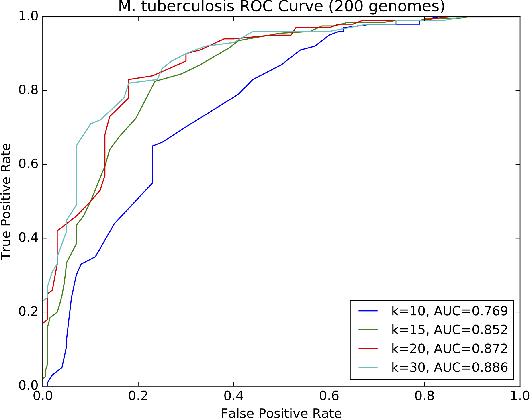
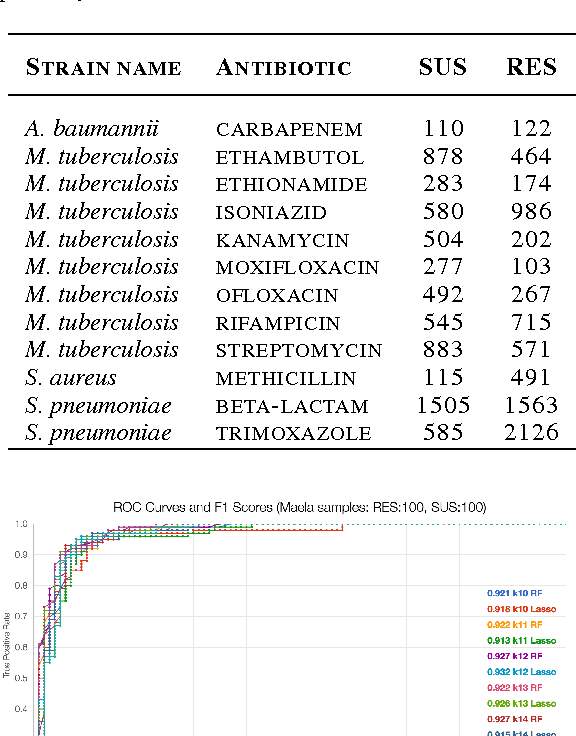
Abstract:Biological datasets amenable to applied machine learning are more available today than ever before, yet they lack adequate representation in the Data-for-Good community. Here we present a work in progress case study performing analysis on antimicrobial resistance (AMR) using standard ensemble machine learning techniques and note the successes and pitfalls such work entails. Broadly, applied machine learning (AML) techniques are well suited to AMR, with classification accuracies ranging from mid-90% to low- 80% depending on sample size. Additionally, these techniques prove successful at identifying gene regions known to be associated with the AMR phenotype. We believe that the extensive amount of biological data available, the plethora of problems presented, and the global impact of such work merits the consideration of the Data- for-Good community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge