Dragan Pamucar
IRIT-ADRIA, IRIT, UT1
Unified Review and Benchmark of Deep Segmentation Architectures for Cardiac Ultrasound on CAMUS
Dec 27, 2025Abstract:Several review papers summarize cardiac imaging and DL advances, few works connect this overview to a unified and reproducible experimental benchmark. In this study, we combine a focused review of cardiac ultrasound segmentation literature with a controlled comparison of three influential architectures, U-Net, Attention U-Net, and TransUNet, on the Cardiac Acquisitions for Multi-Structure Ultrasound Segmentation (CAMUS) echocardiography dataset. Our benchmark spans multiple preprocessing routes, including native NIfTI volumes, 16-bit PNG exports, GPT-assisted polygon-based pseudo-labels, and self-supervised pretraining (SSL) on thousands of unlabeled cine frames. Using identical training splits, losses, and evaluation criteria, a plain U-Net achieved a 94% mean Dice when trained directly on NIfTI data (preserving native dynamic range), while the PNG-16-bit workflow reached 91% under similar conditions. Attention U-Net provided modest improvements on small or low-contrast regions, reducing boundary leakage, whereas TransUNet demonstrated the strongest generalization on challenging frames due to its ability to model global spatial context, particularly when initialized with SSL. Pseudo-labeling expanded the training set and improved robustness after confidence filtering. Overall, our contributions are threefold: a harmonized, apples-to-apples benchmark of U-Net, Attention U-Net, and TransUNet under standardized CAMUS preprocessing and evaluation; practical guidance on maintaining intensity fidelity, resolution consistency, and alignment when preparing ultrasound data; and an outlook on scalable self-supervision and emerging multimodal GPT-based annotation pipelines for rapid labeling, quality assurance, and targeted dataset curation.
Hybrid Ensemble Approaches: Optimal Deep Feature Fusion and Hyperparameter-Tuned Classifier Ensembling for Enhanced Brain Tumor Classification
Jul 16, 2025Abstract:Magnetic Resonance Imaging (MRI) is widely recognized as the most reliable tool for detecting tumors due to its capability to produce detailed images that reveal their presence. However, the accuracy of diagnosis can be compromised when human specialists evaluate these images. Factors such as fatigue, limited expertise, and insufficient image detail can lead to errors. For example, small tumors might go unnoticed, or overlap with healthy brain regions could result in misidentification. To address these challenges and enhance diagnostic precision, this study proposes a novel double ensembling framework, consisting of ensembled pre-trained deep learning (DL) models for feature extraction and ensembled fine-tuned hyperparameter machine learning (ML) models to efficiently classify brain tumors. Specifically, our method includes extensive preprocessing and augmentation, transfer learning concepts by utilizing various pre-trained deep convolutional neural networks and vision transformer networks to extract deep features from brain MRI, and fine-tune hyperparameters of ML classifiers. Our experiments utilized three different publicly available Kaggle MRI brain tumor datasets to evaluate the pre-trained DL feature extractor models, ML classifiers, and the effectiveness of an ensemble of deep features along with an ensemble of ML classifiers for brain tumor classification. Our results indicate that the proposed feature fusion and classifier fusion improve upon the state of the art, with hyperparameter fine-tuning providing a significant enhancement over the ensemble method. Additionally, we present an ablation study to illustrate how each component contributes to accurate brain tumor classification.
A VIKOR and TOPSIS focused reanalysis of the MADM methods based on logarithmic normalization
Jun 15, 2020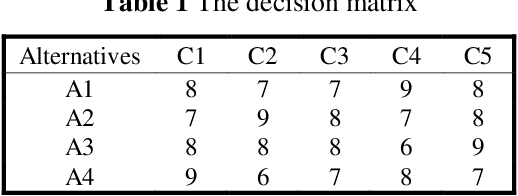
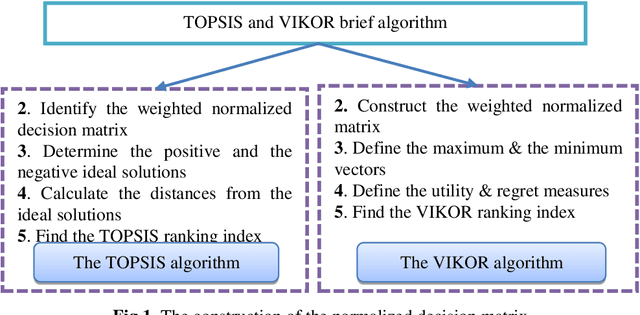
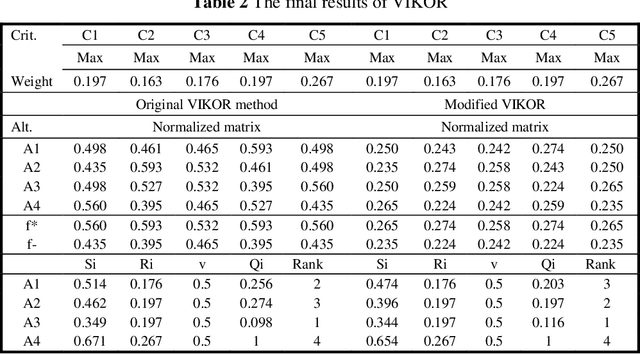
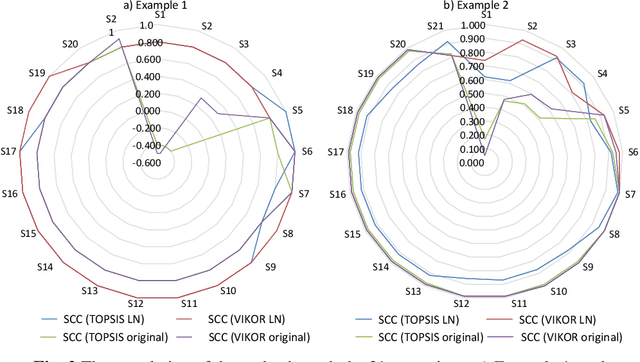
Abstract:Decision and policy-makers in multi-criteria decision-making analysis take into account some strategies in order to analyze outcomes and to finally make an effective and more precise decision. Among those strategies, the modification of the normalization process in the multiple-criteria decision-making algorithm is still a question due to the confrontation of many normalization tools. Normalization is the basic action in defining and solving a MADM problem and a MADM model. Normalization is the first, also necessary, step in solving, i.e. the application of a MADM method. It is a fact that the selection of normalization methods has a direct effect on the results. One of the latest normalization methods introduced is the Logarithmic Normalization (LN) method. This new method has a distinguished advantage, reflecting in that a sum of the normalized values of criteria always equals 1. This normalization method had never been applied in any MADM methods before. This research study is focused on the analysis of the classical MADM methods based on logarithmic normalization. VIKOR and TOPSIS, as the two famous MADM methods, were selected for this reanalysis research study. Two numerical examples were checked in both methods, based on both the classical and the novel ways based on the LN. The results indicate that there are differences between the two approaches. Eventually, a sensitivity analysis is also designed to illustrate the reliability of the final results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge