David Van Valen
Department of Biology, California Institute of Technology, Pasadena, CA, USA
Adaptive Protein Tokenization
Feb 06, 2026Abstract:Tokenization is a promising path to multi-modal models capable of jointly understanding protein sequences, structure, and function. Existing protein structure tokenizers create tokens by pooling information from local neighborhoods, an approach that limits their performance on generative and representation tasks. In this work, we present a method for global tokenization of protein structures in which successive tokens contribute increasing levels of detail to a global representation. This change resolves several issues with generative models based on local protein tokenization: it mitigates error accumulation, provides embeddings without sequence-reduction operations, and allows task-specific adaptation of a tokenized sequence's information content. We validate our method on reconstruction, generative, and representation tasks and demonstrate that it matches or outperforms existing models based on local protein structure tokenizers. We show how adaptive tokens enable inference criteria based on information content, which boosts designability. We validate representations generated from our tokenizer on CATH classification tasks and demonstrate that non-linear probing on our tokenized sequences outperforms equivalent probing on representations from other tokenizers. Finally, we demonstrate how our method supports zero-shot protein shrinking and affinity maturation.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021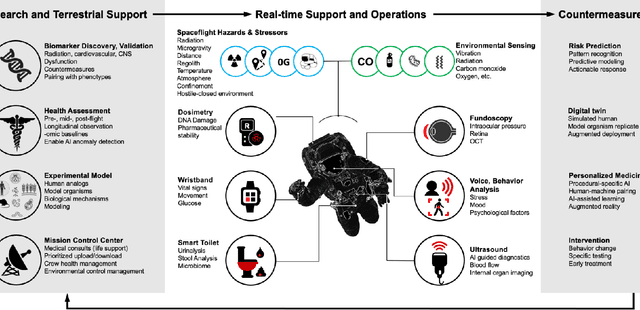
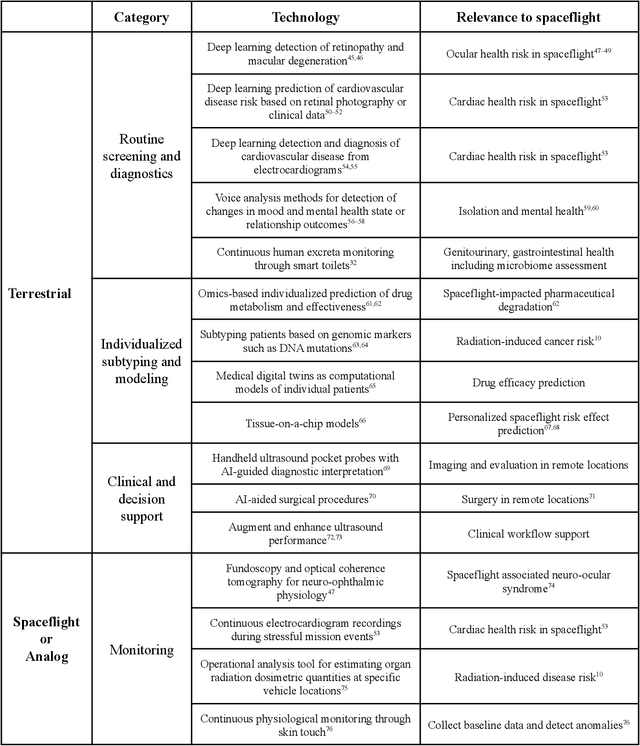
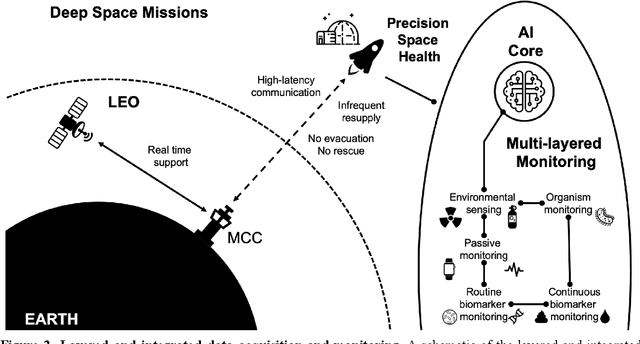
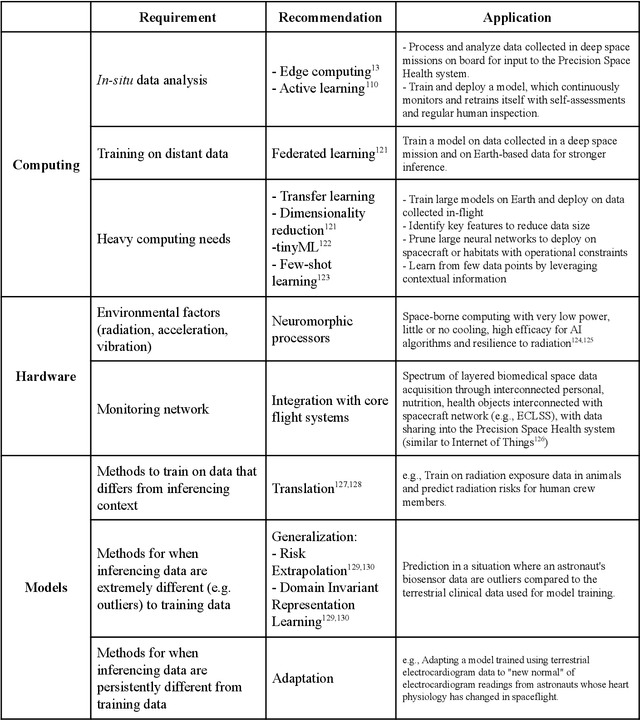
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
Beyond Low Earth Orbit: Biological Research, Artificial Intelligence, and Self-Driving Labs
Dec 22, 2021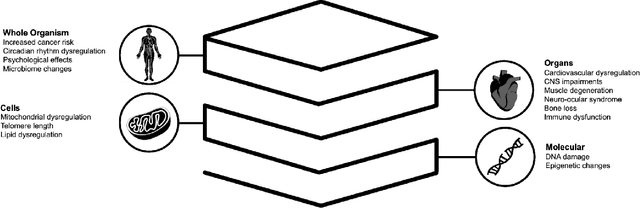
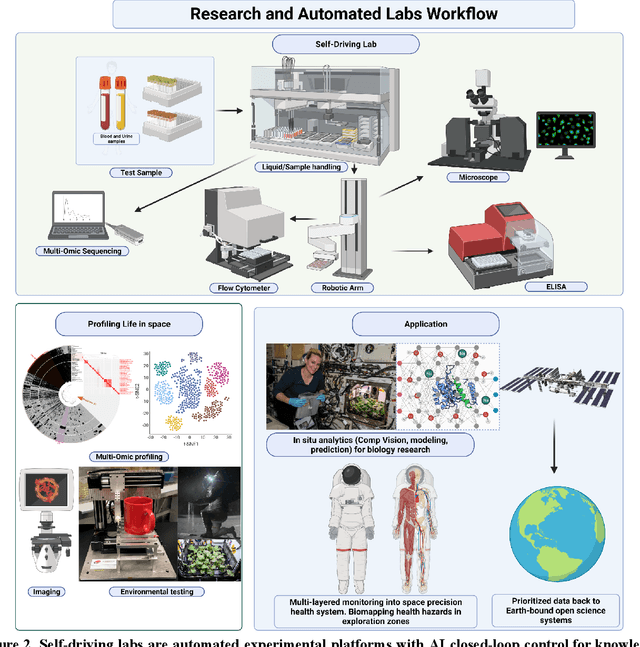
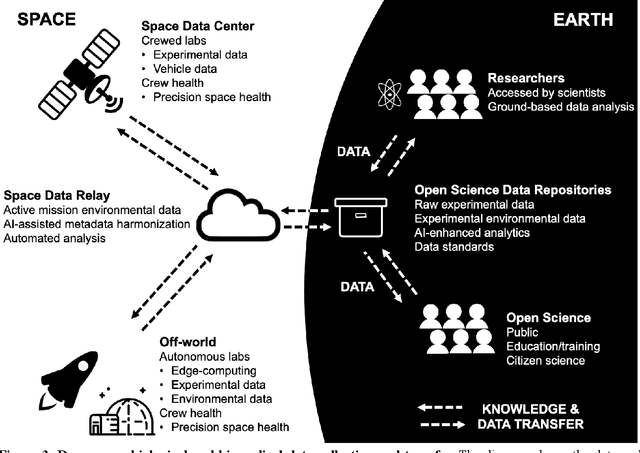
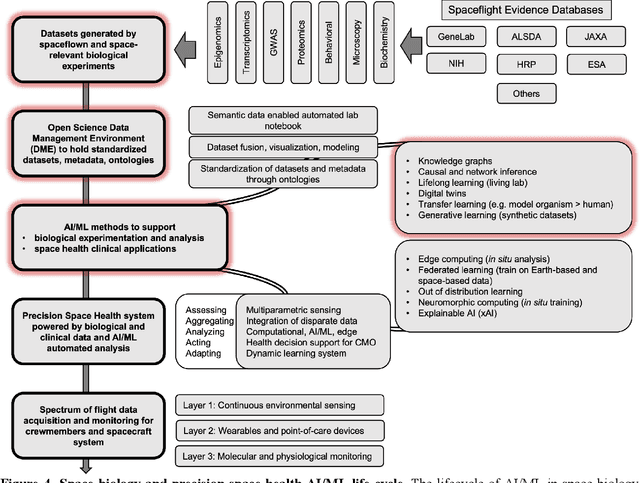
Abstract:Space biology research aims to understand fundamental effects of spaceflight on organisms, develop foundational knowledge to support deep space exploration, and ultimately bioengineer spacecraft and habitats to stabilize the ecosystem of plants, crops, microbes, animals, and humans for sustained multi-planetary life. To advance these aims, the field leverages experiments, platforms, data, and model organisms from both spaceborne and ground-analog studies. As research is extended beyond low Earth orbit, experiments and platforms must be maximally autonomous, light, agile, and intelligent to expedite knowledge discovery. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration on artificial intelligence, machine learning, and modeling applications which offer key solutions toward these space biology challenges. In the next decade, the synthesis of artificial intelligence into the field of space biology will deepen the biological understanding of spaceflight effects, facilitate predictive modeling and analytics, support maximally autonomous and reproducible experiments, and efficiently manage spaceborne data and metadata, all with the goal to enable life to thrive in deep space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge