Ciaran Bench
Investigation into using stochastic embedding representations for evaluating the trustworthiness of the Fréchet Inception Distance
Jan 29, 2026Abstract:Feature embeddings acquired from pretrained models are widely used in medical applications of deep learning to assess the characteristics of datasets; e.g. to determine the quality of synthetic, generated medical images. The Fréchet Inception Distance (FID) is one popular synthetic image quality metric that relies on the assumption that the characteristic features of the data can be detected and encoded by an InceptionV3 model pretrained on ImageNet1K (natural images). While it is widely known that this makes it less effective for applications involving medical images, the extent to which the metric fails to capture meaningful differences in image characteristics is not obviously known. Here, we use Monte Carlo dropout to compute the predictive variance in the FID as well as a supplemental estimate of the predictive variance in the feature embedding model's latent representations. We show that the magnitudes of the predictive variances considered exhibit varying degrees of correlation with the extent to which test inputs (ImageNet1K validation set augmented at various strengths, and other external datasets) are out-of-distribution relative to its training data, providing some insight into the effectiveness of their use as indicators of the trustworthiness of the FID.
Uncertainty quantification with approximate variational learning for wearable photoplethysmography prediction tasks
May 16, 2025Abstract:Photoplethysmography (PPG) signals encode information about relative changes in blood volume that can be used to assess various aspects of cardiac health non-invasively, e.g.\ to detect atrial fibrillation (AF) or predict blood pressure (BP). Deep networks are well-equipped to handle the large quantities of data acquired from wearable measurement devices. However, they lack interpretability and are prone to overfitting, leaving considerable risk for poor performance on unseen data and misdiagnosis. Here, we describe the use of two scalable uncertainty quantification techniques: Monte Carlo Dropout and the recently proposed Improved Variational Online Newton. These techniques are used to assess the trustworthiness of models trained to perform AF classification and BP regression from raw PPG time series. We find that the choice of hyperparameters has a considerable effect on the predictive performance of the models and on the quality and composition of predicted uncertainties. E.g. the stochasticity of the model parameter sampling determines the proportion of the total uncertainty that is aleatoric, and has varying effects on predictive performance and calibration quality dependent on the chosen uncertainty quantification technique and the chosen expression of uncertainty. We find significant discrepancy in the quality of uncertainties over the predicted classes, emphasising the need for a thorough evaluation protocol that assesses local and adaptive calibration. This work suggests that the choice of hyperparameters must be carefully tuned to balance predictive performance and calibration quality, and that the optimal parameterisation may vary depending on the chosen expression of uncertainty.
Quantifying the uncertainty of model-based synthetic image quality metrics
Apr 04, 2025Abstract:The quality of synthetically generated images (e.g. those produced by diffusion models) are often evaluated using information about image contents encoded by pretrained auxiliary models. For example, the Fr\'{e}chet Inception Distance (FID) uses embeddings from an InceptionV3 model pretrained to classify ImageNet. The effectiveness of this feature embedding model has considerable impact on the trustworthiness of the calculated metric (affecting its suitability in several domains, including medical imaging). Here, uncertainty quantification (UQ) is used to provide a heuristic measure of the trustworthiness of the feature embedding model and an FID-like metric called the Fr\'{e}chet Autoencoder Distance (FAED). We apply Monte Carlo dropout to a feature embedding model (convolutional autoencoder) to model the uncertainty in its embeddings. The distribution of embeddings for each input are then used to compute a distribution of FAED values. We express uncertainty as the predictive variance of the embeddings as well as the standard deviation of the computed FAED values. We find that their magnitude correlates with the extent to which the inputs are out-of-distribution to the model's training data, providing some validation of its ability to assess the trustworthiness of the FAED.
Machine-learning for photoplethysmography analysis: Benchmarking feature, image, and signal-based approaches
Feb 27, 2025



Abstract:Photoplethysmography (PPG) is a widely used non-invasive physiological sensing technique, suitable for various clinical applications. Such clinical applications are increasingly supported by machine learning methods, raising the question of the most appropriate input representation and model choice. Comprehensive comparisons, in particular across different input representations, are scarce. We address this gap in the research landscape by a comprehensive benchmarking study covering three kinds of input representations, interpretable features, image representations and raw waveforms, across prototypical regression and classification use cases: blood pressure and atrial fibrillation prediction. In both cases, the best results are achieved by deep neural networks operating on raw time series as input representations. Within this model class, best results are achieved by modern convolutional neural networks (CNNs). but depending on the task setup, shallow CNNs are often also very competitive. We envision that these results will be insightful for researchers to guide their choice on machine learning tasks for PPG data, even beyond the use cases presented in this work.
Style transfer as data augmentation: evaluating unpaired image-to-image translation models in mammography
Feb 04, 2025



Abstract:Several studies indicate that deep learning models can learn to detect breast cancer from mammograms (X-ray images of the breasts). However, challenges with overfitting and poor generalisability prevent their routine use in the clinic. Models trained on data from one patient population may not perform well on another due to differences in their data domains, emerging due to variations in scanning technology or patient characteristics. Data augmentation techniques can be used to improve generalisability by expanding the diversity of feature representations in the training data by altering existing examples. Image-to-image translation models are one approach capable of imposing the characteristic feature representations (i.e. style) of images from one dataset onto another. However, evaluating model performance is non-trivial, particularly in the absence of ground truths (a common reality in medical imaging). Here, we describe some key aspects that should be considered when evaluating style transfer algorithms, highlighting the advantages and disadvantages of popular metrics, and important factors to be mindful of when implementing them in practice. We consider two types of generative models: a cycle-consistent generative adversarial network (CycleGAN) and a diffusion-based SynDiff model. We learn unpaired image-to-image translation across three mammography datasets. We highlight that undesirable aspects of model performance may determine the suitability of some metrics, and also provide some analysis indicating the extent to which various metrics assess unique aspects of model performance. We emphasise the need to use several metrics for a comprehensive assessment of model performance.
Trustworthy image-to-image translation: evaluating uncertainty calibration in unpaired training scenarios
Jan 29, 2025



Abstract:Mammographic screening is an effective method for detecting breast cancer, facilitating early diagnosis. However, the current need to manually inspect images places a heavy burden on healthcare systems, spurring a desire for automated diagnostic protocols. Techniques based on deep neural networks have been shown effective in some studies, but their tendency to overfit leaves considerable risk for poor generalisation and misdiagnosis, preventing their widespread adoption in clinical settings. Data augmentation schemes based on unpaired neural style transfer models have been proposed that improve generalisability by diversifying the representations of training image features in the absence of paired training data (images of the same tissue in either image style). But these models are similarly prone to various pathologies, and evaluating their performance is challenging without ground truths/large datasets (as is often the case in medical imaging). Here, we consider two frameworks/architectures: a GAN-based cycleGAN, and the more recently developed diffusion-based SynDiff. We evaluate their performance when trained on image patches parsed from three open access mammography datasets and one non-medical image dataset. We consider the use of uncertainty quantification to assess model trustworthiness, and propose a scheme to evaluate calibration quality in unpaired training scenarios. This ultimately helps facilitate the trustworthy use of image-to-image translation models in domains where ground truths are not typically available.
Unsupervised segmentation of biomedical hyperspectral image data: tackling high dimensionality with convolutional autoencoders
Sep 09, 2022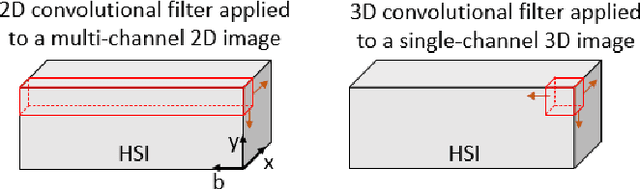
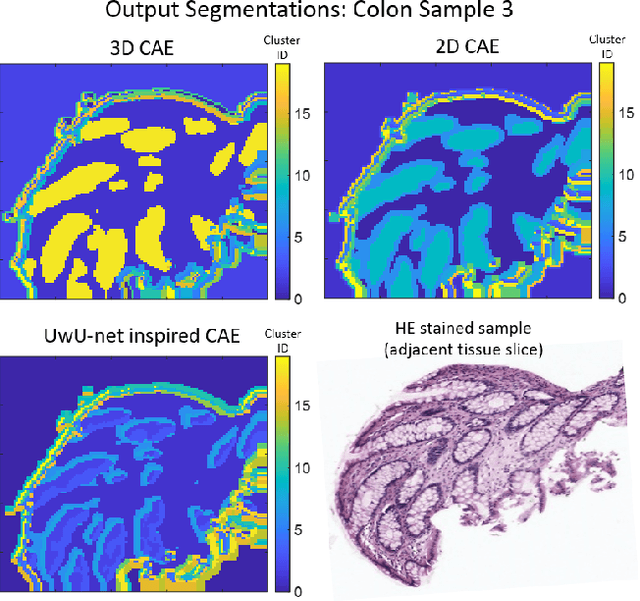
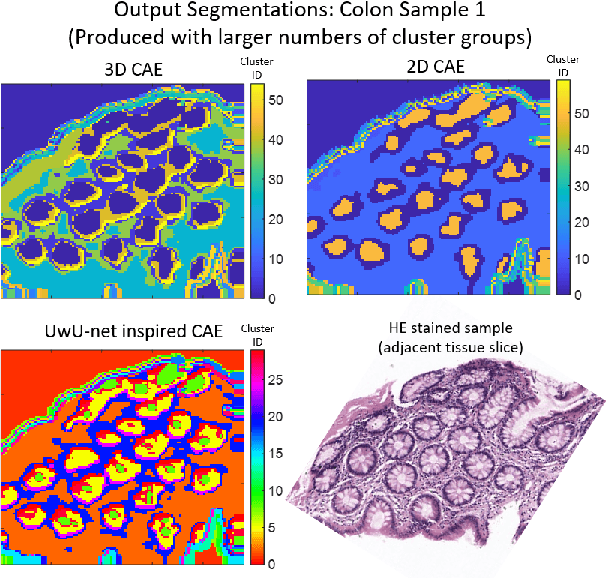
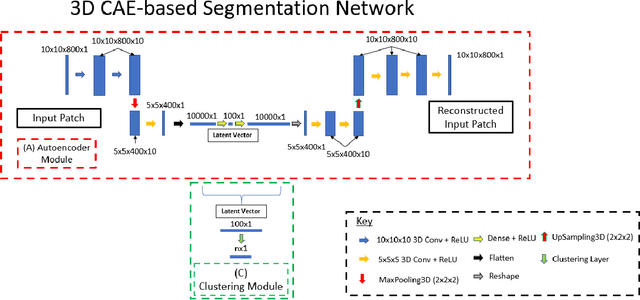
Abstract:Information about the structure and composition of biopsy specimens can assist in disease monitoring and diagnosis. In principle, this can be acquired from Raman and infrared (IR) hyperspectral images (HSIs) that encode information about how a sample's constituent molecules are arranged in space. Each tissue section/component is defined by a unique combination of spatial and spectral features, but given the high dimensionality of HSI datasets, extracting and utilising them to segment images is non-trivial. Here, we show how networks based on deep convolutional autoencoders (CAEs) can perform this task in an end-to-end fashion by first detecting and compressing relevant features from patches of the HSI into low-dimensional latent vectors, and then performing a clustering step that groups patches containing similar spatio-spectral features together. We showcase the advantages of using this end-to-end spatio-spectral segmentation approach compared to i) the same spatio-spectral technique not trained in an end-to-end manner, and ii) a method that only utilises spectral features (spectral k-means) using simulated HSIs of porcine tissue as test examples. Secondly, we describe the potential advantages/limitations of using three different CAE architectures: a generic 2D CAE, a generic 3D CAE, and a 2D CNN architecture inspired by the recently proposed UwU-net that is specialised for extracting features from HSI data. We assess their performance on IR HSIs of real colon samples. We find that all architectures are capable of producing segmentations that show good correspondence with HE stained adjacent tissue slices used as approximate ground truths, indicating the robustness of the CAE-driven approach for segmenting biomedical HSI data. Additionally, we stress the need for more accurate ground truth information to rigorously compare the advantages offered by each architecture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge