Spencer A. Thomas
Investigation into using stochastic embedding representations for evaluating the trustworthiness of the Fréchet Inception Distance
Jan 29, 2026Abstract:Feature embeddings acquired from pretrained models are widely used in medical applications of deep learning to assess the characteristics of datasets; e.g. to determine the quality of synthetic, generated medical images. The Fréchet Inception Distance (FID) is one popular synthetic image quality metric that relies on the assumption that the characteristic features of the data can be detected and encoded by an InceptionV3 model pretrained on ImageNet1K (natural images). While it is widely known that this makes it less effective for applications involving medical images, the extent to which the metric fails to capture meaningful differences in image characteristics is not obviously known. Here, we use Monte Carlo dropout to compute the predictive variance in the FID as well as a supplemental estimate of the predictive variance in the feature embedding model's latent representations. We show that the magnitudes of the predictive variances considered exhibit varying degrees of correlation with the extent to which test inputs (ImageNet1K validation set augmented at various strengths, and other external datasets) are out-of-distribution relative to its training data, providing some insight into the effectiveness of their use as indicators of the trustworthiness of the FID.
Quantifying the uncertainty of model-based synthetic image quality metrics
Apr 04, 2025Abstract:The quality of synthetically generated images (e.g. those produced by diffusion models) are often evaluated using information about image contents encoded by pretrained auxiliary models. For example, the Fr\'{e}chet Inception Distance (FID) uses embeddings from an InceptionV3 model pretrained to classify ImageNet. The effectiveness of this feature embedding model has considerable impact on the trustworthiness of the calculated metric (affecting its suitability in several domains, including medical imaging). Here, uncertainty quantification (UQ) is used to provide a heuristic measure of the trustworthiness of the feature embedding model and an FID-like metric called the Fr\'{e}chet Autoencoder Distance (FAED). We apply Monte Carlo dropout to a feature embedding model (convolutional autoencoder) to model the uncertainty in its embeddings. The distribution of embeddings for each input are then used to compute a distribution of FAED values. We express uncertainty as the predictive variance of the embeddings as well as the standard deviation of the computed FAED values. We find that their magnitude correlates with the extent to which the inputs are out-of-distribution to the model's training data, providing some validation of its ability to assess the trustworthiness of the FAED.
Style transfer as data augmentation: evaluating unpaired image-to-image translation models in mammography
Feb 04, 2025



Abstract:Several studies indicate that deep learning models can learn to detect breast cancer from mammograms (X-ray images of the breasts). However, challenges with overfitting and poor generalisability prevent their routine use in the clinic. Models trained on data from one patient population may not perform well on another due to differences in their data domains, emerging due to variations in scanning technology or patient characteristics. Data augmentation techniques can be used to improve generalisability by expanding the diversity of feature representations in the training data by altering existing examples. Image-to-image translation models are one approach capable of imposing the characteristic feature representations (i.e. style) of images from one dataset onto another. However, evaluating model performance is non-trivial, particularly in the absence of ground truths (a common reality in medical imaging). Here, we describe some key aspects that should be considered when evaluating style transfer algorithms, highlighting the advantages and disadvantages of popular metrics, and important factors to be mindful of when implementing them in practice. We consider two types of generative models: a cycle-consistent generative adversarial network (CycleGAN) and a diffusion-based SynDiff model. We learn unpaired image-to-image translation across three mammography datasets. We highlight that undesirable aspects of model performance may determine the suitability of some metrics, and also provide some analysis indicating the extent to which various metrics assess unique aspects of model performance. We emphasise the need to use several metrics for a comprehensive assessment of model performance.
Trustworthy image-to-image translation: evaluating uncertainty calibration in unpaired training scenarios
Jan 29, 2025



Abstract:Mammographic screening is an effective method for detecting breast cancer, facilitating early diagnosis. However, the current need to manually inspect images places a heavy burden on healthcare systems, spurring a desire for automated diagnostic protocols. Techniques based on deep neural networks have been shown effective in some studies, but their tendency to overfit leaves considerable risk for poor generalisation and misdiagnosis, preventing their widespread adoption in clinical settings. Data augmentation schemes based on unpaired neural style transfer models have been proposed that improve generalisability by diversifying the representations of training image features in the absence of paired training data (images of the same tissue in either image style). But these models are similarly prone to various pathologies, and evaluating their performance is challenging without ground truths/large datasets (as is often the case in medical imaging). Here, we consider two frameworks/architectures: a GAN-based cycleGAN, and the more recently developed diffusion-based SynDiff. We evaluate their performance when trained on image patches parsed from three open access mammography datasets and one non-medical image dataset. We consider the use of uncertainty quantification to assess model trustworthiness, and propose a scheme to evaluate calibration quality in unpaired training scenarios. This ultimately helps facilitate the trustworthy use of image-to-image translation models in domains where ground truths are not typically available.
Cluster Metric Sensitivity to Irrelevant Features
Feb 19, 2024


Abstract:Clustering algorithms are used extensively in data analysis for data exploration and discovery. Technological advancements lead to continually growth of data in terms of volume, dimensionality and complexity. This provides great opportunities in data analytics as the data can be interrogated for many different purposes. This however leads challenges, such as identification of relevant features for a given task. In supervised tasks, one can utilise a number of methods to optimise the input features for the task objective (e.g. classification accuracy). In unsupervised problems, such tools are not readily available, in part due to an inability to quantify feature relevance in unlabeled tasks. In this paper, we investigate the sensitivity of clustering performance noisy uncorrelated variables iteratively added to baseline datasets with well defined clusters. We show how different types of irrelevant variables can impact the outcome of a clustering result from $k$-means in different ways. We observe a resilience to very high proportions of irrelevant features for adjusted rand index (ARI) and normalised mutual information (NMI) when the irrelevant features are Gaussian distributed. For Uniformly distributed irrelevant features, we notice the resilience of ARI and NMI is dependent on the dimensionality of the data and exhibits tipping points between high scores and near zero. Our results show that the Silhouette Coefficient and the Davies-Bouldin score are the most sensitive to irrelevant added features exhibiting large changes in score for comparably low proportions of irrelevant features regardless of underlying distribution or data scaling. As such the Silhouette Coefficient and the Davies-Bouldin score are good candidates for optimising feature selection in unsupervised clustering tasks.
Fast Data Driven Estimation of Cluster Number in Multiplex Images using Embedded Density Outliers
Jul 21, 2022



Abstract:The usage of chemical imaging technologies is becoming a routine accompaniment to traditional methods in pathology. Significant technological advances have developed these next generation techniques to provide rich, spatially resolved, multidimensional chemical images. The rise of digital pathology has significantly enhanced the synergy of these imaging modalities with optical microscopy and immunohistochemistry, enhancing our understanding of the biological mechanisms and progression of diseases. Techniques such as imaging mass cytometry provide labelled multidimensional (multiplex) images of specific components used in conjunction with digital pathology techniques. These powerful techniques generate a wealth of high dimensional data that create significant challenges in data analysis. Unsupervised methods such as clustering are an attractive way to analyse these data, however, they require the selection of parameters such as the number of clusters. Here we propose a methodology to estimate the number of clusters in an automatic data-driven manner using a deep sparse autoencoder to embed the data into a lower dimensional space. We compute the density of regions in the embedded space, the majority of which are empty, enabling the high density regions to be detected as outliers and provide an estimate for the number of clusters. This framework provides a fully unsupervised and data-driven method to analyse multidimensional data. In this work we demonstrate our method using 45 multiplex imaging mass cytometry datasets. Moreover, our model is trained using only one of the datasets and the learned embedding is applied to the remaining 44 images providing an efficient process for data analysis. Finally, we demonstrate the high computational efficiency of our method which is two orders of magnitude faster than estimating via computing the sum squared distances as a function of cluster number.
A Generic Self-Supervised Framework of Learning Invariant Discriminative Features
Feb 14, 2022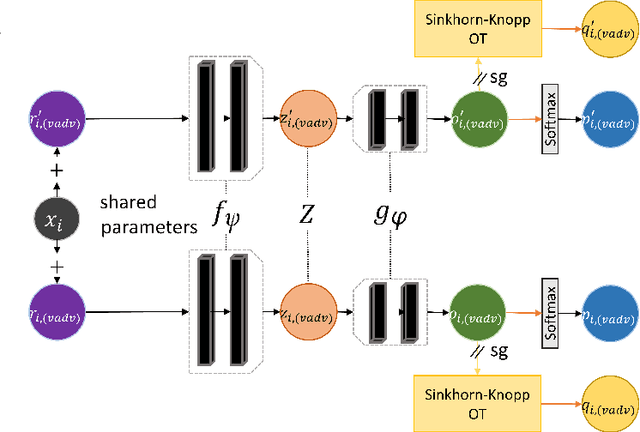
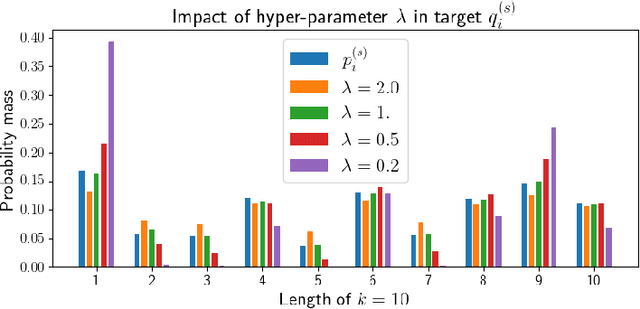
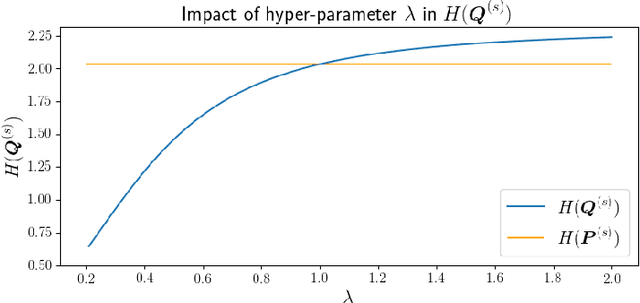
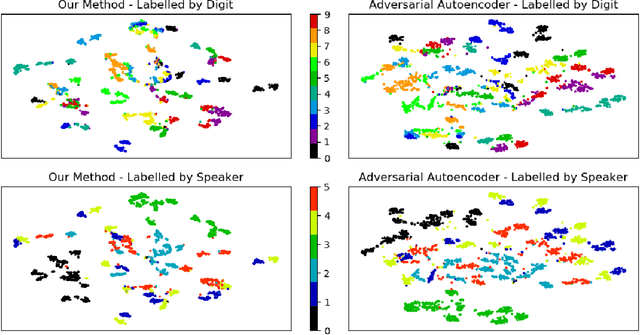
Abstract:Self-supervised learning (SSL) has become a popular method for generating invariant representations without the need for human annotations. Nonetheless, the desired invariant representation is achieved by utilising prior online transformation functions on the input data. As a result, each SSL framework is customised for a particular data type, e.g., visual data, and further modifications are required if it is used for other dataset types. On the other hand, autoencoder (AE), which is a generic and widely applicable framework, mainly focuses on dimension reduction and is not suited for learning invariant representation. This paper proposes a generic SSL framework based on a constrained self-labelling assignment process that prevents degenerate solutions. Specifically, the prior transformation functions are replaced with a self-transformation mechanism, derived through an unsupervised training process of adversarial training, for imposing invariant representations. Via the self-transformation mechanism, pairs of augmented instances can be generated from the same input data. Finally, a training objective based on contrastive learning is designed by leveraging both the self-labelling assignment and the self-transformation mechanism. Despite the fact that the self-transformation process is very generic, the proposed training strategy outperforms a majority of state-of-the-art representation learning methods based on AE structures. To validate the performance of our method, we conduct experiments on four types of data, namely visual, audio, text, and mass spectrometry data, and compare them in terms of four quantitative metrics. Our comparison results indicate that the proposed method demonstrate robustness and successfully identify patterns within the datasets.
Enhanced Transfer Learning Through Medical Imaging and Patient Demographic Data Fusion
Nov 29, 2021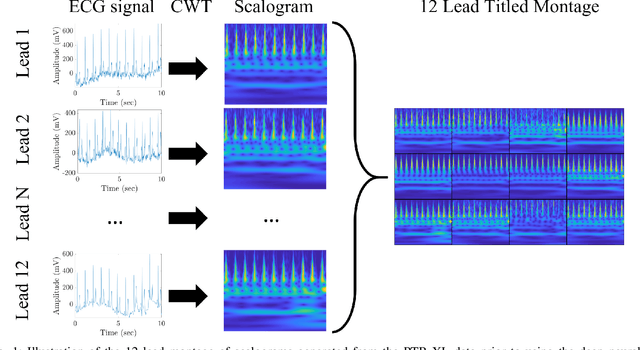
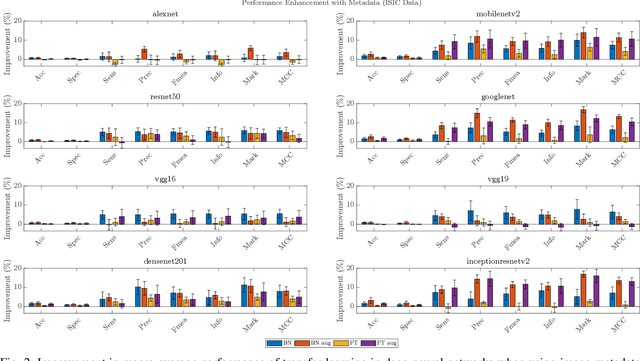
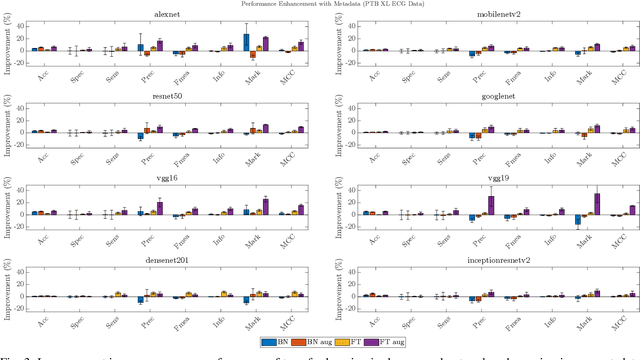
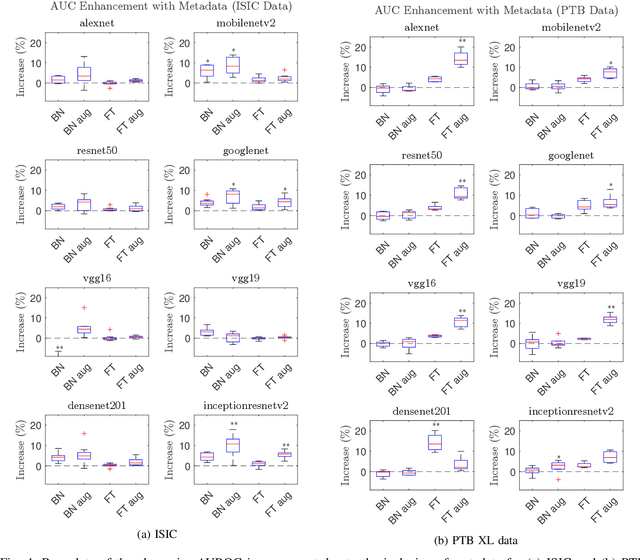
Abstract:In this work we examine the performance enhancement in classification of medical imaging data when image features are combined with associated non-image data. We compare the performance of eight state-of-the-art deep neural networks in classification tasks when using only image features, compared to when these are combined with patient metadata. We utilise transfer learning with networks pretrained on ImageNet used directly as feature extractors and fine tuned on the target domain. Our experiments show that performance can be significantly enhanced with the inclusion of metadata and use interpretability methods to identify which features lead to these enhancements. Furthermore, our results indicate that the performance enhancement for natural medical imaging (e.g. optical images) benefit most from direct use of pre-trained models, whereas non natural images (e.g. representations of non imaging data) benefit most from fine tuning pre-trained networks. These enhancements come at a negligible additional cost in computation time, and therefore is a practical method for other applications.
Combining Image Features and Patient Metadata to Enhance Transfer Learning
Oct 08, 2021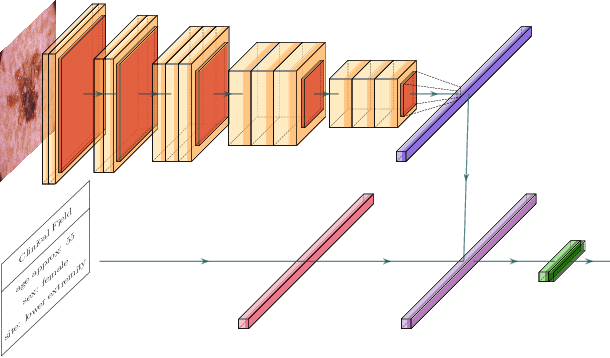
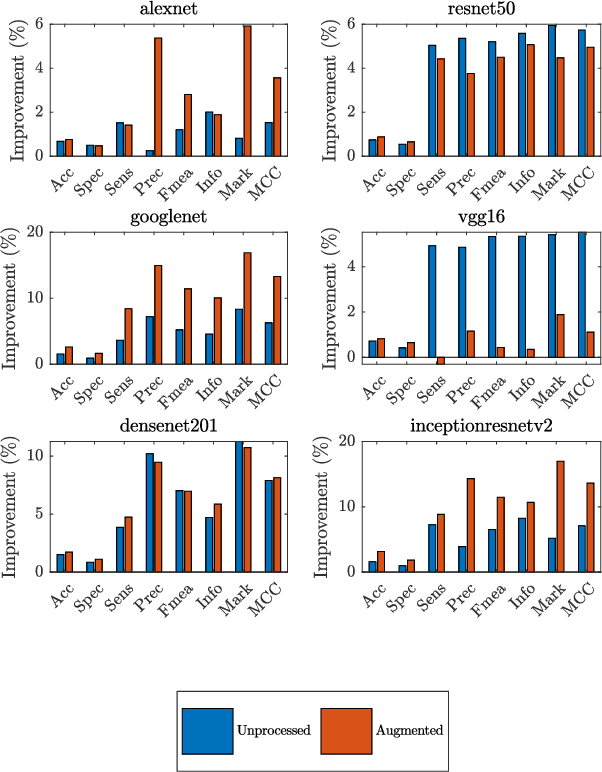
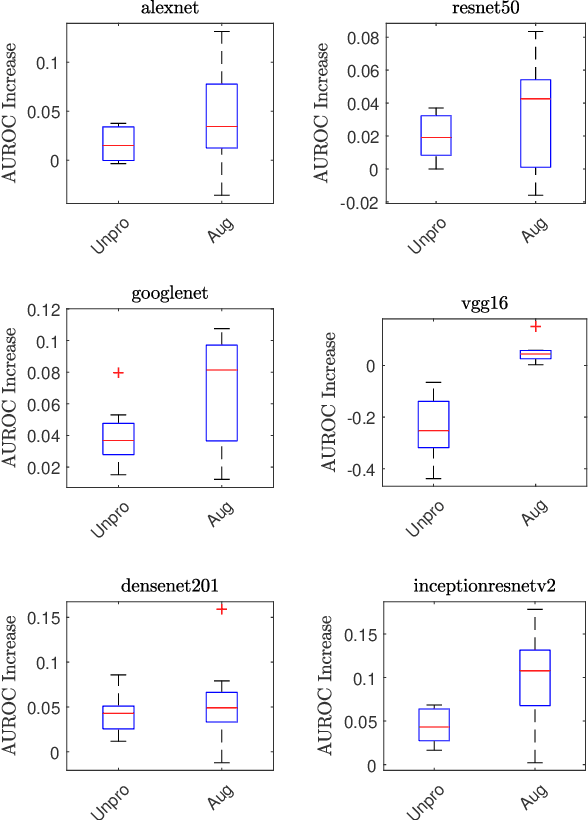
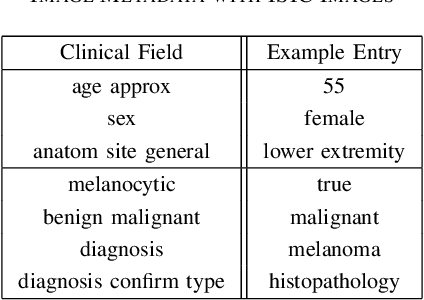
Abstract:In this work, we compare the performance of six state-of-the-art deep neural networks in classification tasks when using only image features, to when these are combined with patient metadata. We utilise transfer learning from networks pretrained on ImageNet to extract image features from the ISIC HAM10000 dataset prior to classification. Using several classification performance metrics, we evaluate the effects of including metadata with the image features. Furthermore, we repeat our experiments with data augmentation. Our results show an overall enhancement in performance of each network as assessed by all metrics, only noting degradation in a vgg16 architecture. Our results indicate that this performance enhancement may be a general property of deep networks and should be explored in other areas. Moreover, these improvements come at a negligible additional cost in computation time, and therefore are a practical method for other applications.
Information Maximization Clustering via Multi-View Self-Labelling
Mar 12, 2021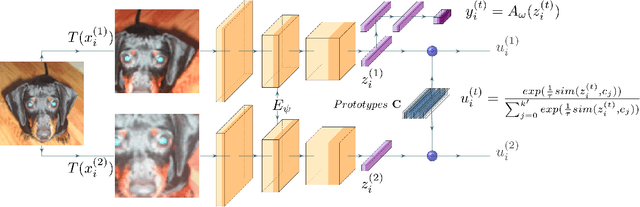
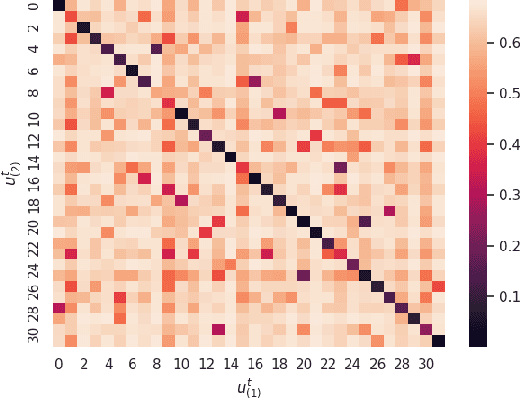
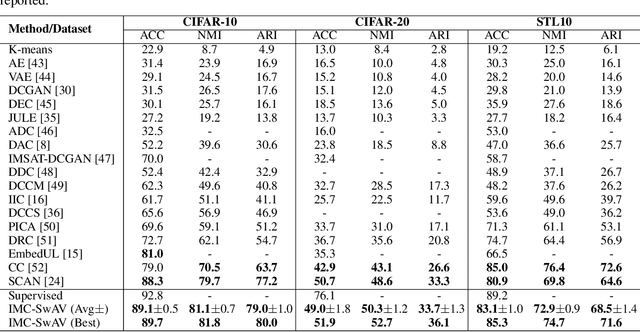
Abstract:Image clustering is a particularly challenging computer vision task, which aims to generate annotations without human supervision. Recent advances focus on the use of self-supervised learning strategies in image clustering, by first learning valuable semantics and then clustering the image representations. These multiple-phase algorithms, however, increase the computational time and their final performance is reliant on the first stage. By extending the self-supervised approach, we propose a novel single-phase clustering method that simultaneously learns meaningful representations and assigns the corresponding annotations. This is achieved by integrating a discrete representation into the self-supervised paradigm through a classifier net. Specifically, the proposed clustering objective employs mutual information, and maximizes the dependency between the integrated discrete representation and a discrete probability distribution. The discrete probability distribution is derived though the self-supervised process by comparing the learnt latent representation with a set of trainable prototypes. To enhance the learning performance of the classifier, we jointly apply the mutual information across multi-crop views. Our empirical results show that the proposed framework outperforms state-of-the-art techniques with the average accuracy of 89.1% and 49.0%, respectively, on CIFAR-10 and CIFAR-100/20 datasets. Finally, the proposed method also demonstrates attractive robustness to parameter settings, making it ready to be applicable to other datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge