Chuanjie Wu
When to use Graphs in RAG: A Comprehensive Analysis for Graph Retrieval-Augmented Generation
Jun 06, 2025Abstract:Graph retrieval-augmented generation (GraphRAG) has emerged as a powerful paradigm for enhancing large language models (LLMs) with external knowledge. It leverages graphs to model the hierarchical structure between specific concepts, enabling more coherent and effective knowledge retrieval for accurate reasoning.Despite its conceptual promise, recent studies report that GraphRAG frequently underperforms vanilla RAG on many real-world tasks. This raises a critical question: Is GraphRAG really effective, and in which scenarios do graph structures provide measurable benefits for RAG systems? To address this, we propose GraphRAG-Bench, a comprehensive benchmark designed to evaluate GraphRAG models onboth hierarchical knowledge retrieval and deep contextual reasoning. GraphRAG-Bench features a comprehensive dataset with tasks of increasing difficulty, coveringfact retrieval, complex reasoning, contextual summarization, and creative generation, and a systematic evaluation across the entire pipeline, from graph constructionand knowledge retrieval to final generation. Leveraging this novel benchmark, we systematically investigate the conditions when GraphRAG surpasses traditional RAG and the underlying reasons for its success, offering guidelines for its practical application. All related resources and analyses are collected for the community at https://github.com/GraphRAG-Bench/GraphRAG-Benchmark.
Schrödinger-ANI: An Eight-Element Neural Network Interaction Potential with Greatly Expanded Coverage of Druglike Chemical Space
Nov 22, 2019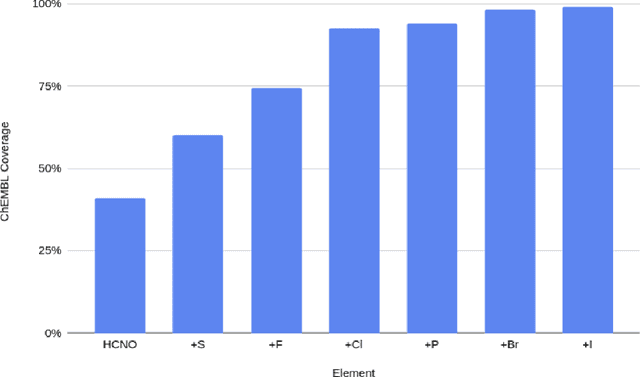
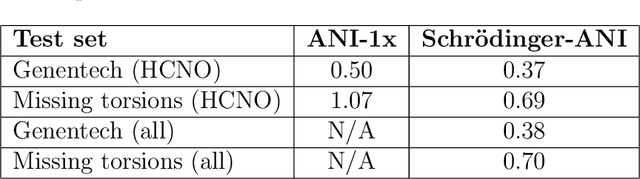
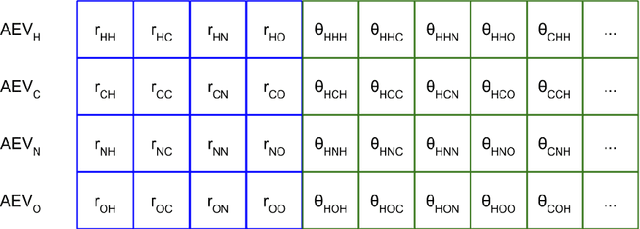
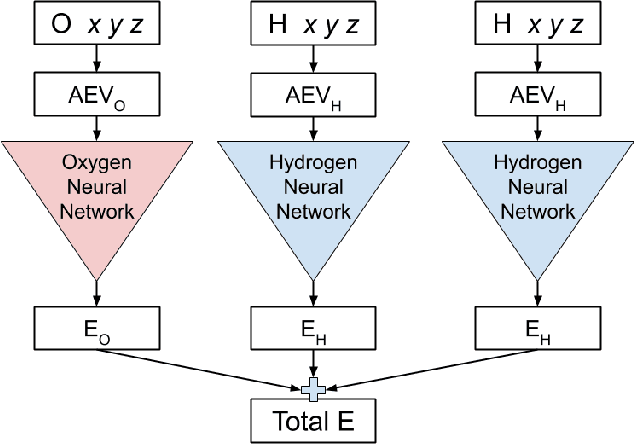
Abstract:We have developed a neural network potential energy function for use in drug discovery, with chemical element support extended from 41% to 94% of druglike molecules based on ChEMBL. We expand on the work of Smith et al., with their highly accurate network for the elements H, C, N, O, creating a network for H, C, N, O, S, F, Cl, P. We focus particularly on the calculation of relative conformer energies, for which we show that our new potential energy function has an RMSE of 0.70 kcal/mol for prospective druglike molecule conformers, substantially better than the previous state of the art. The speed and accuracy of this model could greatly accelerate the parameterization of protein-ligand binding free energy calculations for novel druglike molecules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge