Christine Klymko
VERIRAG: Healthcare Claim Verification via Statistical Audit in Retrieval-Augmented Generation
Jul 23, 2025Abstract:Retrieval-augmented generation (RAG) systems are increasingly adopted in clinical decision support, yet they remain methodologically blind-they retrieve evidence but cannot vet its scientific quality. A paper claiming "Antioxidant proteins decreased after alloferon treatment" and a rigorous multi-laboratory replication study will be treated as equally credible, even if the former lacked scientific rigor or was even retracted. To address this challenge, we introduce VERIRAG, a framework that makes three notable contributions: (i) the Veritable, an 11-point checklist that evaluates each source for methodological rigor, including data integrity and statistical validity; (ii) a Hard-to-Vary (HV) Score, a quantitative aggregator that weights evidence by its quality and diversity; and (iii) a Dynamic Acceptance Threshold, which calibrates the required evidence based on how extraordinary a claim is. Across four datasets-comprising retracted, conflicting, comprehensive, and settled science corpora-the VERIRAG approach consistently outperforms all baselines, achieving absolute F1 scores ranging from 0.53 to 0.65, representing a 10 to 14 point improvement over the next-best method in each respective dataset. We will release all materials necessary for reproducing our results.
An Ensemble Framework for Detecting Community Changes in Dynamic Networks
Jul 24, 2017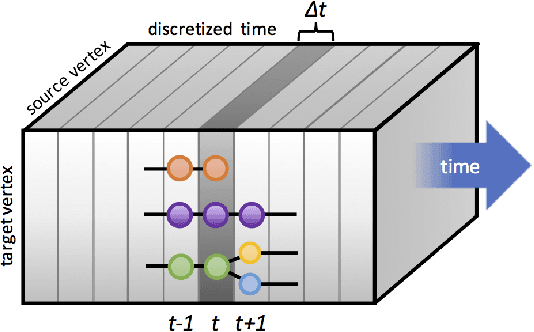
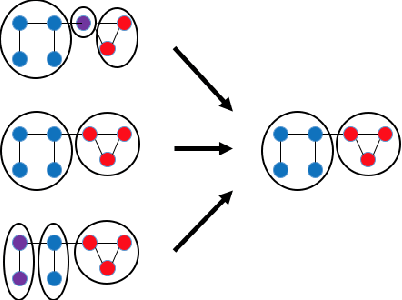
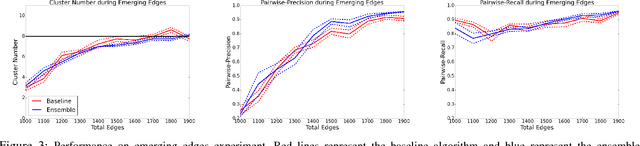
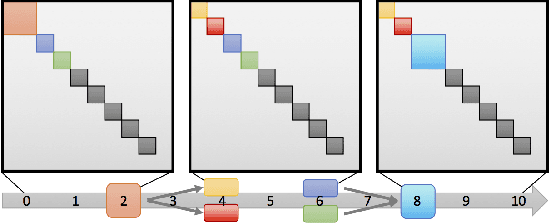
Abstract:Dynamic networks, especially those representing social networks, undergo constant evolution of their community structure over time. Nodes can migrate between different communities, communities can split into multiple new communities, communities can merge together, etc. In order to represent dynamic networks with evolving communities it is essential to use a dynamic model rather than a static one. Here we use a dynamic stochastic block model where the underlying block model is different at different times. In order to represent the structural changes expressed by this dynamic model the network will be split into discrete time segments and a clustering algorithm will assign block memberships for each segment. In this paper we show that using an ensemble of clustering assignments accommodates for the variance in scalable clustering algorithms and produces superior results in terms of pairwise-precision and pairwise-recall. We also demonstrate that the dynamic clustering produced by the ensemble can be visualized as a flowchart which encapsulates the community evolution succinctly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge