Chencheng Xu
Phi-Former: A Pairwise Hierarchical Approach for Compound-Protein Interactions Prediction
Feb 05, 2026Abstract:Drug discovery remains time-consuming, labor-intensive, and expensive, often requiring years and substantial investment per drug candidate. Predicting compound-protein interactions (CPIs) is a critical component in this process, enabling the identification of molecular interactions between drug candidates and target proteins. Recent deep learning methods have successfully modeled CPIs at the atomic level, achieving improved efficiency and accuracy over traditional energy-based approaches. However, these models do not always align with chemical realities, as molecular fragments (motifs or functional groups) typically serve as the primary units of biological recognition and binding. In this paper, we propose Phi-former, a pairwise hierarchical interaction representation learning method that addresses this gap by incorporating the biological role of motifs in CPIs. Phi-former represents compounds and proteins hierarchically and employs a pairwise pre-training framework to model interactions systematically across atom-atom, motif-motif, and atom-motif levels, reflecting how biological systems recognize molecular partners. We design intra-level and inter-level learning pipelines that make different interaction levels mutually beneficial. Experimental results demonstrate that Phi-former achieves superior performance on CPI-related tasks. A case study shows that our method accurately identifies specific atoms or motifs activated in CPIs, providing interpretable model explanations. These insights may guide rational drug design and support precision medicine applications.
CFP-Gen: Combinatorial Functional Protein Generation via Diffusion Language Models
May 28, 2025Abstract:Existing PLMs generate protein sequences based on a single-condition constraint from a specific modality, struggling to simultaneously satisfy multiple constraints across different modalities. In this work, we introduce CFP-Gen, a novel diffusion language model for Combinatorial Functional Protein GENeration. CFP-Gen facilitates the de novo protein design by integrating multimodal conditions with functional, sequence, and structural constraints. Specifically, an Annotation-Guided Feature Modulation (AGFM) module is introduced to dynamically adjust the protein feature distribution based on composable functional annotations, e.g., GO terms, IPR domains and EC numbers. Meanwhile, the Residue-Controlled Functional Encoding (RCFE) module captures residue-wise interaction to ensure more precise control. Additionally, off-the-shelf 3D structure encoders can be seamlessly integrated to impose geometric constraints. We demonstrate that CFP-Gen enables high-throughput generation of novel proteins with functionality comparable to natural proteins, while achieving a high success rate in designing multifunctional proteins. Code and data available at https://github.com/yinjunbo/cfpgen.
Acceleration of Federated Learning with Alleviated Forgetting in Local Training
Mar 05, 2022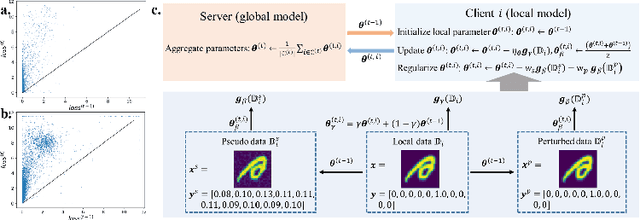
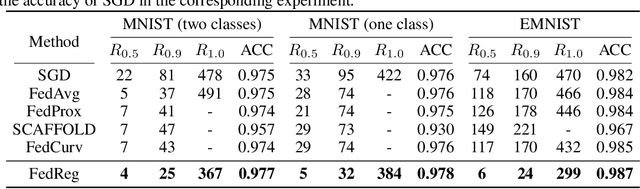
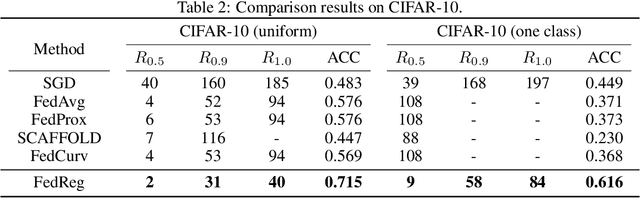
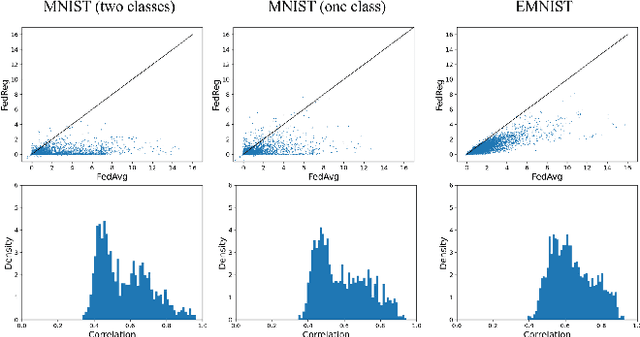
Abstract:Federated learning (FL) enables distributed optimization of machine learning models while protecting privacy by independently training local models on each client and then aggregating parameters on a central server, thereby producing an effective global model. Although a variety of FL algorithms have been proposed, their training efficiency remains low when the data are not independently and identically distributed (non-i.i.d.) across different clients. We observe that the slow convergence rates of the existing methods are (at least partially) caused by the catastrophic forgetting issue during the local training stage on each individual client, which leads to a large increase in the loss function concerning the previous training data at the other clients. Here, we propose FedReg, an algorithm to accelerate FL with alleviated knowledge forgetting in the local training stage by regularizing locally trained parameters with the loss on generated pseudo data, which encode the knowledge of previous training data learned by the global model. Our comprehensive experiments demonstrate that FedReg not only significantly improves the convergence rate of FL, especially when the neural network architecture is deep and the clients' data are extremely non-i.i.d., but is also able to protect privacy better in classification problems and more robust against gradient inversion attacks. The code is available at: https://github.com/Zoesgithub/FedReg.
Reinforced Molecular Optimization with Neighborhood-Controlled Grammars
Nov 14, 2020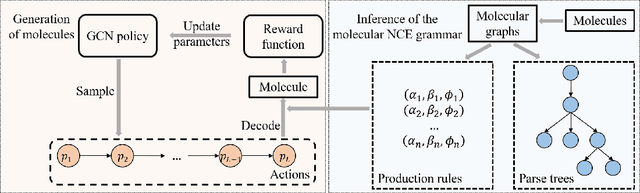
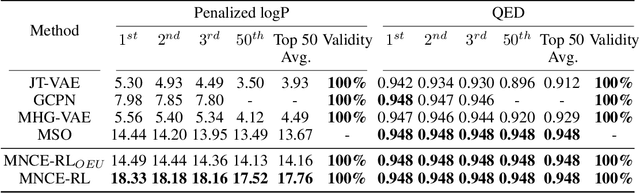
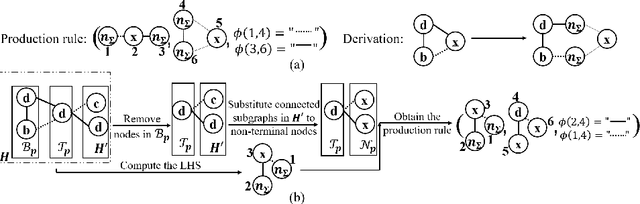
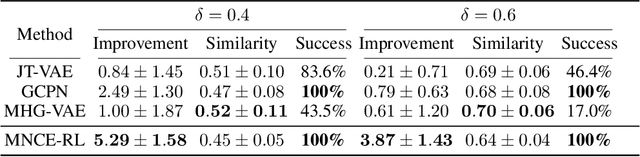
Abstract:A major challenge in the pharmaceutical industry is to design novel molecules with specific desired properties, especially when the property evaluation is costly. Here, we propose MNCE-RL, a graph convolutional policy network for molecular optimization with molecular neighborhood-controlled embedding grammars through reinforcement learning. We extend the original neighborhood-controlled embedding grammars to make them applicable to molecular graph generation and design an efficient algorithm to infer grammatical production rules from given molecules. The use of grammars guarantees the validity of the generated molecular structures. By transforming molecular graphs to parse trees with the inferred grammars, the molecular structure generation task is modeled as a Markov decision process where a policy gradient strategy is utilized. In a series of experiments, we demonstrate that our approach achieves state-of-the-art performance in a diverse range of molecular optimization tasks and exhibits significant superiority in optimizing molecular properties with a limited number of property evaluations.
* 12 pages;two figures; NeurIPS 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge