Charlotta Lindvall
BioClinical ModernBERT: A State-of-the-Art Long-Context Encoder for Biomedical and Clinical NLP
Jun 12, 2025Abstract:Encoder-based transformer models are central to biomedical and clinical Natural Language Processing (NLP), as their bidirectional self-attention makes them well-suited for efficiently extracting structured information from unstructured text through discriminative tasks. However, encoders have seen slower development compared to decoder models, leading to limited domain adaptation in biomedical and clinical settings. We introduce BioClinical ModernBERT, a domain-adapted encoder that builds on the recent ModernBERT release, incorporating long-context processing and substantial improvements in speed and performance for biomedical and clinical NLP. BioClinical ModernBERT is developed through continued pretraining on the largest biomedical and clinical corpus to date, with over 53.5 billion tokens, and addresses a key limitation of prior clinical encoders by leveraging 20 datasets from diverse institutions, domains, and geographic regions, rather than relying on data from a single source. It outperforms existing biomedical and clinical encoders on four downstream tasks spanning a broad range of use cases. We release both base (150M parameters) and large (396M parameters) versions of BioClinical ModernBERT, along with training checkpoints to support further research.
MedSlice: Fine-Tuned Large Language Models for Secure Clinical Note Sectioning
Jan 23, 2025Abstract:Extracting sections from clinical notes is crucial for downstream analysis but is challenging due to variability in formatting and labor-intensive nature of manual sectioning. While proprietary large language models (LLMs) have shown promise, privacy concerns limit their accessibility. This study develops a pipeline for automated note sectioning using open-source LLMs, focusing on three sections: History of Present Illness, Interval History, and Assessment and Plan. We fine-tuned three open-source LLMs to extract sections using a curated dataset of 487 progress notes, comparing results relative to proprietary models (GPT-4o, GPT-4o mini). Internal and external validity were assessed via precision, recall and F1 score. Fine-tuned Llama 3.1 8B outperformed GPT-4o (F1=0.92). On the external validity test set, performance remained high (F1= 0.85). Fine-tuned open-source LLMs can surpass proprietary models in clinical note sectioning, offering advantages in cost, performance, and accessibility.
Clinical XLNet: Modeling Sequential Clinical Notes and Predicting Prolonged Mechanical Ventilation
Dec 27, 2019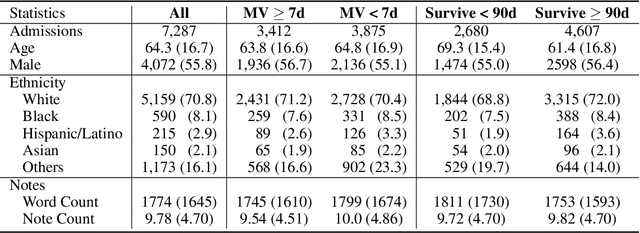
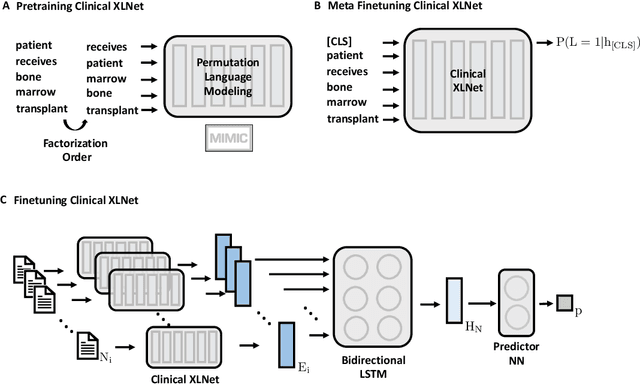
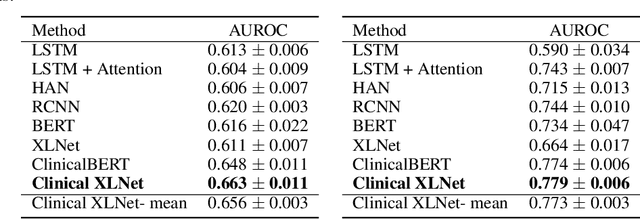
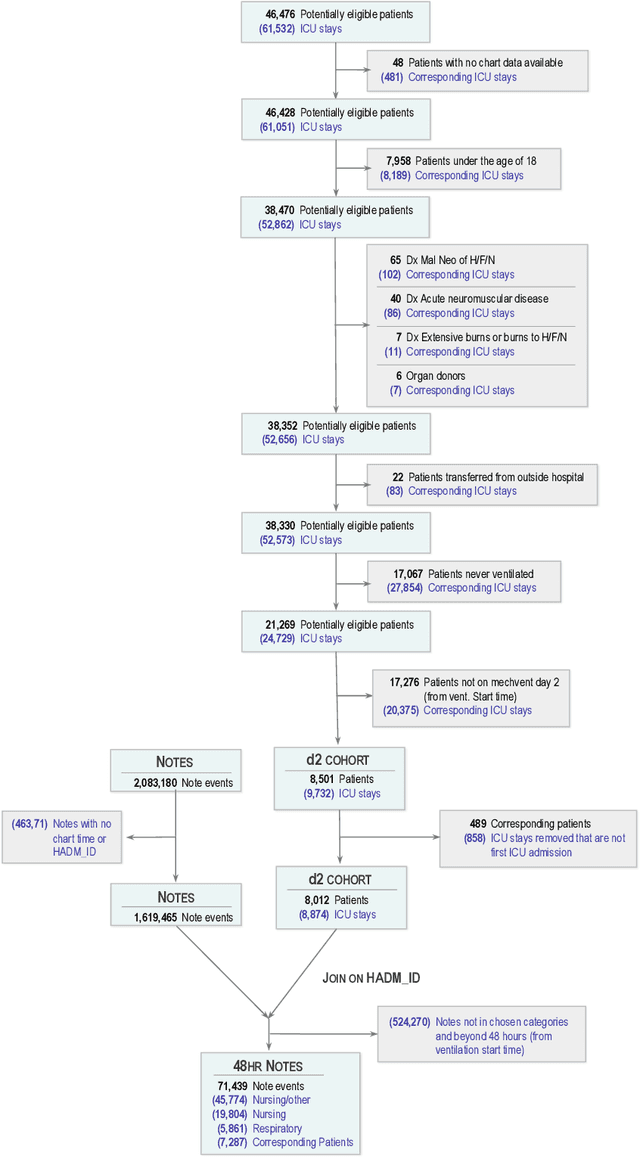
Abstract:Clinical notes contain rich data, which is unexploited in predictive modeling compared to structured data. In this work, we developed a new text representation Clinical XLNet for clinical notes which also leverages the temporal information of the sequence of the notes. We evaluated our models on prolonged mechanical ventilation prediction problem and our experiments demonstrated that Clinical XLNet outperforms the best baselines consistently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge