Tom J. Pollard
BioClinical ModernBERT: A State-of-the-Art Long-Context Encoder for Biomedical and Clinical NLP
Jun 12, 2025Abstract:Encoder-based transformer models are central to biomedical and clinical Natural Language Processing (NLP), as their bidirectional self-attention makes them well-suited for efficiently extracting structured information from unstructured text through discriminative tasks. However, encoders have seen slower development compared to decoder models, leading to limited domain adaptation in biomedical and clinical settings. We introduce BioClinical ModernBERT, a domain-adapted encoder that builds on the recent ModernBERT release, incorporating long-context processing and substantial improvements in speed and performance for biomedical and clinical NLP. BioClinical ModernBERT is developed through continued pretraining on the largest biomedical and clinical corpus to date, with over 53.5 billion tokens, and addresses a key limitation of prior clinical encoders by leveraging 20 datasets from diverse institutions, domains, and geographic regions, rather than relying on data from a single source. It outperforms existing biomedical and clinical encoders on four downstream tasks spanning a broad range of use cases. We release both base (150M parameters) and large (396M parameters) versions of BioClinical ModernBERT, along with training checkpoints to support further research.
MIMIC-CXR: A large publicly available database of labeled chest radiographs
Jan 23, 2019
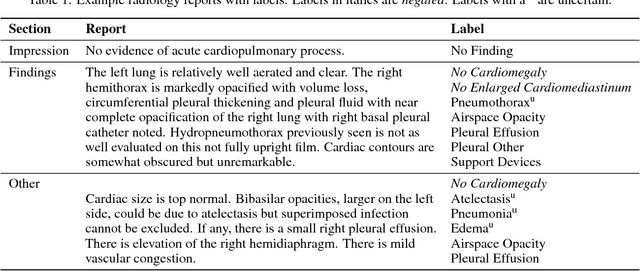
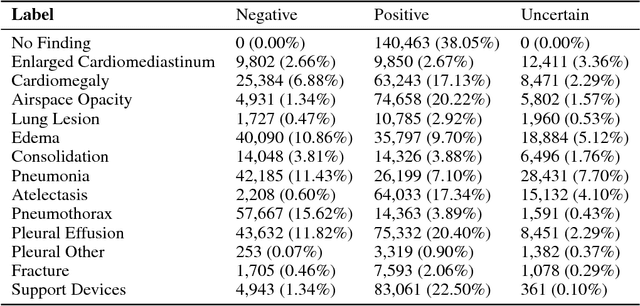
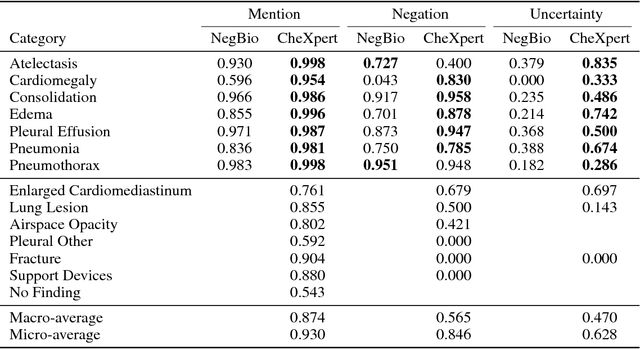
Abstract:Chest radiography is an extremely powerful imaging modality, allowing for a detailed inspection of a patient's thorax, but requiring specialized training for proper interpretation. With the advent of high performance general purpose computer vision algorithms, the accurate automated analysis of chest radiographs is becoming increasingly of interest to researchers. However, a key challenge in the development of these techniques is the lack of sufficient data. Here we describe MIMIC-CXR, a large dataset of 371,920 chest x-rays associated with 227,943 imaging studies sourced from the Beth Israel Deaconess Medical Center between 2011 - 2016. Each imaging study can pertain to one or more images, but most often are associated with two images: a frontal view and a lateral view. Images are provided with 14 labels derived from a natural language processing tool applied to the corresponding free-text radiology reports. All images have been de-identified to protect patient privacy. The dataset is made freely available to facilitate and encourage a wide range of research in medical computer vision.
Generalizability of predictive models for intensive care unit patients
Dec 06, 2018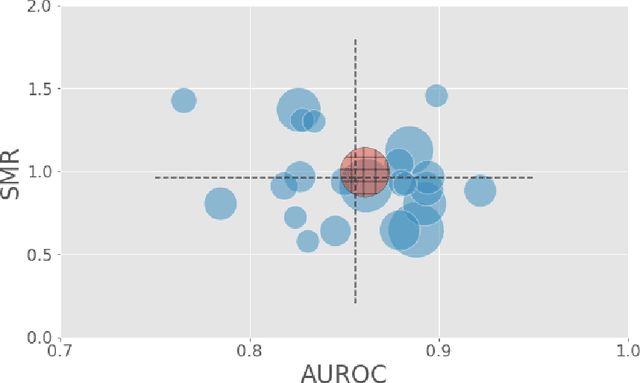
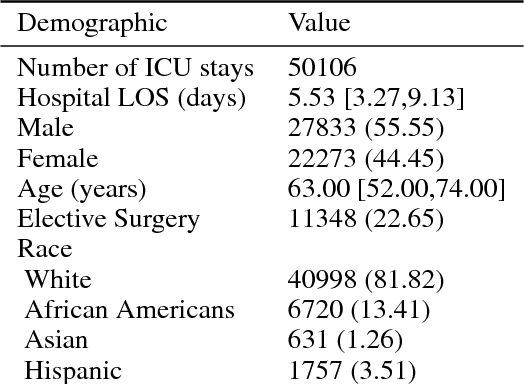
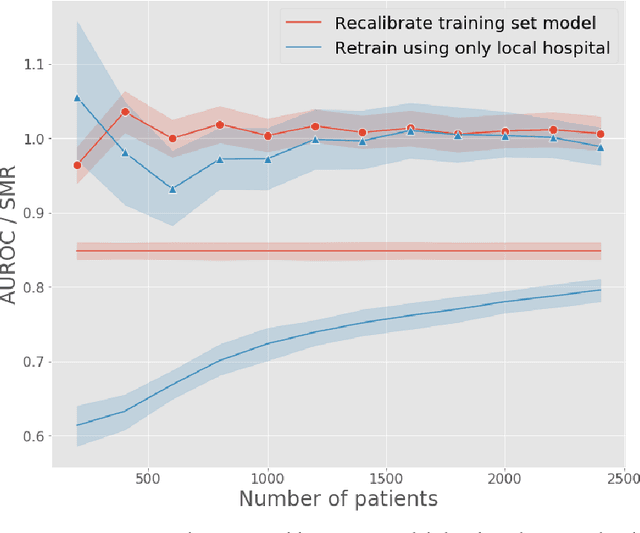
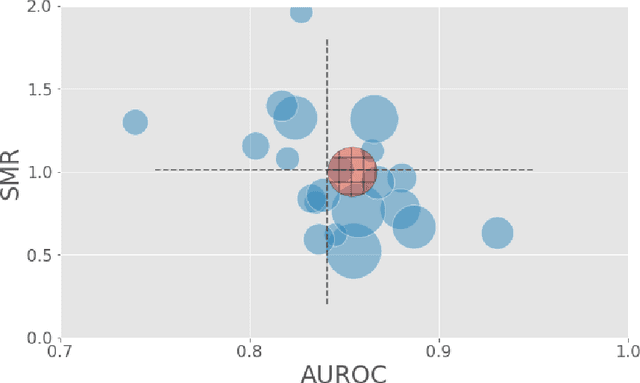
Abstract:A large volume of research has considered the creation of predictive models for clinical data; however, much existing literature reports results using only a single source of data. In this work, we evaluate the performance of models trained on the publicly-available eICU Collaborative Research Database. We show that cross-validation using many distinct centers provides a reasonable estimate of model performance in new centers. We further show that a single model trained across centers transfers well to distinct hospitals, even compared to a model retrained using hospital-specific data. Our results motivate the use of multi-center datasets for model development and highlight the need for data sharing among hospitals to maximize model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge