Chang Min Park
Exploring the Capabilities of LLM Encoders for Image-Text Retrieval in Chest X-rays
Sep 17, 2025



Abstract:Vision-language pretraining has advanced image-text alignment, yet progress in radiology remains constrained by the heterogeneity of clinical reports, including abbreviations, impression-only notes, and stylistic variability. Unlike general-domain settings where more data often leads to better performance, naively scaling to large collections of noisy reports can plateau or even degrade model learning. We ask whether large language model (LLM) encoders can provide robust clinical representations that transfer across diverse styles and better guide image-text alignment. We introduce LLM2VEC4CXR, a domain-adapted LLM encoder for chest X-ray reports, and LLM2CLIP4CXR, a dual-tower framework that couples this encoder with a vision backbone. LLM2VEC4CXR improves clinical text understanding over BERT-based baselines, handles abbreviations and style variation, and achieves strong clinical alignment on report-level metrics. LLM2CLIP4CXR leverages these embeddings to boost retrieval accuracy and clinically oriented scores, with stronger cross-dataset generalization than prior medical CLIP variants. Trained on 1.6M CXR studies from public and private sources with heterogeneous and noisy reports, our models demonstrate that robustness -- not scale alone -- is the key to effective multimodal learning. We release models to support further research in medical image-text representation learning.
AI can evolve without labels: self-evolving vision transformer for chest X-ray diagnosis through knowledge distillation
Feb 13, 2022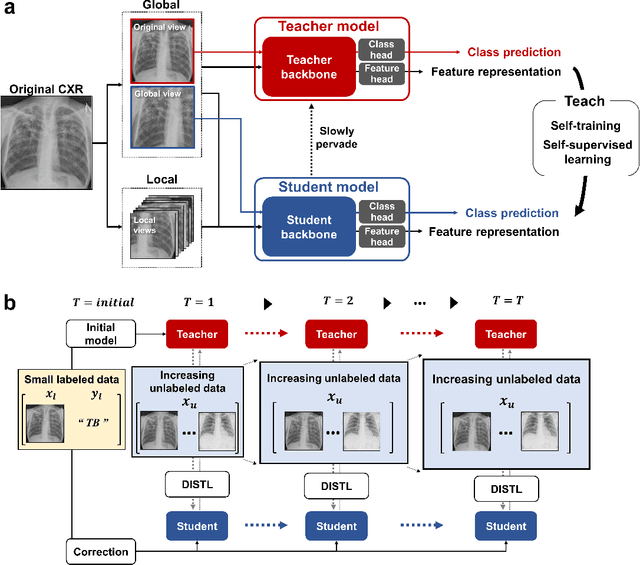
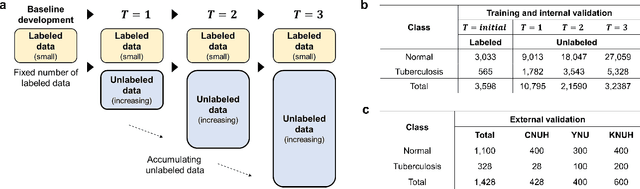
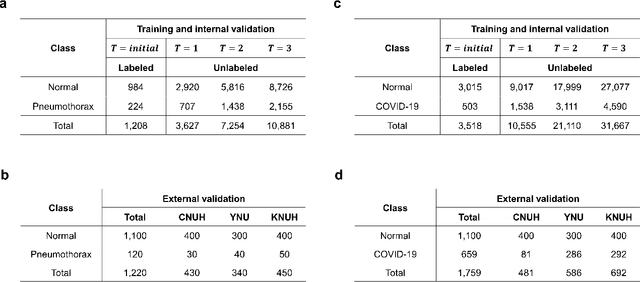
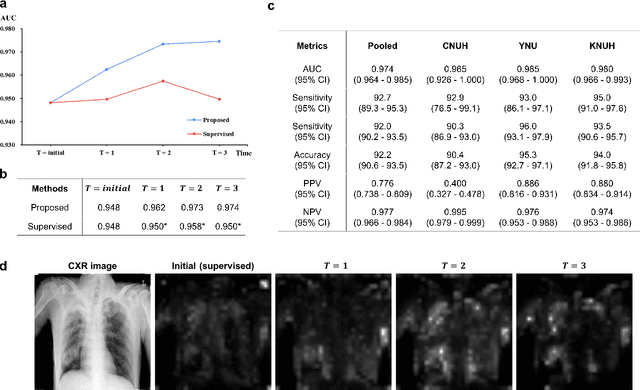
Abstract:Although deep learning-based computer-aided diagnosis systems have recently achieved expert-level performance, developing a robust deep learning model requires large, high-quality data with manual annotation, which is expensive to obtain. This situation poses the problem that the chest x-rays collected annually in hospitals cannot be used due to the lack of manual labeling by experts, especially in deprived areas. To address this, here we present a novel deep learning framework that uses knowledge distillation through self-supervised learning and self-training, which shows that the performance of the original model trained with a small number of labels can be gradually improved with more unlabeled data. Experimental results show that the proposed framework maintains impressive robustness against a real-world environment and has general applicability to several diagnostic tasks such as tuberculosis, pneumothorax, and COVID-19. Notably, we demonstrated that our model performs even better than those trained with the same amount of labeled data. The proposed framework has a great potential for medical imaging, where plenty of data is accumulated every year, but ground truth annotations are expensive to obtain.
Learning Visual Context by Comparison
Jul 15, 2020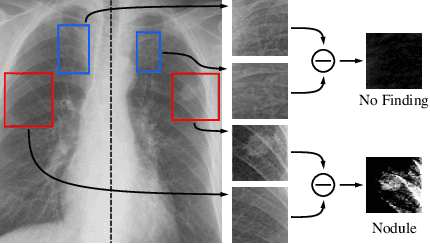
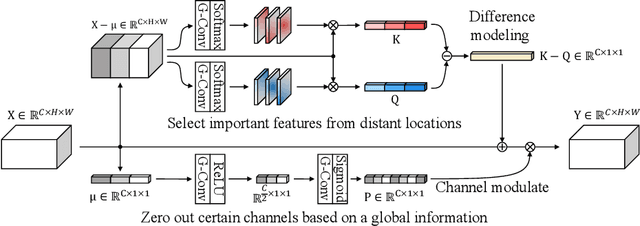
Abstract:Finding diseases from an X-ray image is an important yet highly challenging task. Current methods for solving this task exploit various characteristics of the chest X-ray image, but one of the most important characteristics is still missing: the necessity of comparison between related regions in an image. In this paper, we present Attend-and-Compare Module (ACM) for capturing the difference between an object of interest and its corresponding context. We show that explicit difference modeling can be very helpful in tasks that require direct comparison between locations from afar. This module can be plugged into existing deep learning models. For evaluation, we apply our module to three chest X-ray recognition tasks and COCO object detection & segmentation tasks and observe consistent improvements across tasks. The code is available at https://github.com/mk-minchul/attend-and-compare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge