Chandra Mohan
ConStruct: Structural Distillation of Foundation Models for Prototype-Based Weakly Supervised Histopathology Segmentation
Dec 11, 2025Abstract:Weakly supervised semantic segmentation (WSSS) in histopathology relies heavily on classification backbones, yet these models often localize only the most discriminative regions and struggle to capture the full spatial extent of tissue structures. Vision-language models such as CONCH offer rich semantic alignment and morphology-aware representations, while modern segmentation backbones like SegFormer preserve fine-grained spatial cues. However, combining these complementary strengths remains challenging, especially under weak supervision and without dense annotations. We propose a prototype learning framework for WSSS in histopathological images that integrates morphology-aware representations from CONCH, multi-scale structural cues from SegFormer, and text-guided semantic alignment to produce prototypes that are simultaneously semantically discriminative and spatially coherent. To effectively leverage these heterogeneous sources, we introduce text-guided prototype initialization that incorporates pathology descriptions to generate more complete and semantically accurate pseudo-masks. A structural distillation mechanism transfers spatial knowledge from SegFormer to preserve fine-grained morphological patterns and local tissue boundaries during prototype learning. Our approach produces high-quality pseudo masks without pixel-level annotations, improves localization completeness, and enhances semantic consistency across tissue types. Experiments on BCSS-WSSS datasets demonstrate that our prototype learning framework outperforms existing WSSS methods while remaining computationally efficient through frozen foundation model backbones and lightweight trainable adapters.
DualProtoSeg: Simple and Efficient Design with Text- and Image-Guided Prototype Learning for Weakly Supervised Histopathology Image Segmentation
Dec 11, 2025Abstract:Weakly supervised semantic segmentation (WSSS) in histopathology seeks to reduce annotation cost by learning from image-level labels, yet it remains limited by inter-class homogeneity, intra-class heterogeneity, and the region-shrinkage effect of CAM-based supervision. We propose a simple and effective prototype-driven framework that leverages vision-language alignment to improve region discovery under weak supervision. Our method integrates CoOp-style learnable prompt tuning to generate text-based prototypes and combines them with learnable image prototypes, forming a dual-modal prototype bank that captures both semantic and appearance cues. To address oversmoothing in ViT representations, we incorporate a multi-scale pyramid module that enhances spatial precision and improves localization quality. Experiments on the BCSS-WSSS benchmark show that our approach surpasses existing state-of-the-art methods, and detailed analyses demonstrate the benefits of text description diversity, context length, and the complementary behavior of text and image prototypes. These results highlight the effectiveness of jointly leveraging textual semantics and visual prototype learning for WSSS in digital pathology.
Contrastive Integrated Gradients: A Feature Attribution-Based Method for Explaining Whole Slide Image Classification
Nov 14, 2025Abstract:Interpretability is essential in Whole Slide Image (WSI) analysis for computational pathology, where understanding model predictions helps build trust in AI-assisted diagnostics. While Integrated Gradients (IG) and related attribution methods have shown promise, applying them directly to WSIs introduces challenges due to their high-resolution nature. These methods capture model decision patterns but may overlook class-discriminative signals that are crucial for distinguishing between tumor subtypes. In this work, we introduce Contrastive Integrated Gradients (CIG), a novel attribution method that enhances interpretability by computing contrastive gradients in logit space. First, CIG highlights class-discriminative regions by comparing feature importance relative to a reference class, offering sharper differentiation between tumor and non-tumor areas. Second, CIG satisfies the axioms of integrated attribution, ensuring consistency and theoretical soundness. Third, we propose two attribution quality metrics, MIL-AIC and MIL-SIC, which measure how predictive information and model confidence evolve with access to salient regions, particularly under weak supervision. We validate CIG across three datasets spanning distinct cancer types: CAMELYON16 (breast cancer metastasis in lymph nodes), TCGA-RCC (renal cell carcinoma), and TCGA-Lung (lung cancer). Experimental results demonstrate that CIG yields more informative attributions both quantitatively, using MIL-AIC and MIL-SIC, and qualitatively, through visualizations that align closely with ground truth tumor regions, underscoring its potential for interpretable and trustworthy WSI-based diagnostics
Segmentation of diagnostic tissue compartments on whole slide images with renal thrombotic microangiopathies (TMAs)
Nov 28, 2023



Abstract:The thrombotic microangiopathies (TMAs) manifest in renal biopsy histology with a broad spectrum of acute and chronic findings. Precise diagnostic criteria for a renal biopsy diagnosis of TMA are missing. As a first step towards a machine learning- and computer vision-based analysis of wholes slide images from renal biopsies, we trained a segmentation model for the decisive diagnostic kidney tissue compartments artery, arteriole, glomerulus on a set of whole slide images from renal biopsies with TMAs and Mimickers (distinct diseases with a similar nephropathological appearance as TMA like severe benign nephrosclerosis, various vasculitides, Bevacizumab-plug glomerulopathy, arteriolar light chain deposition disease). Our segmentation model combines a U-Net-based tissue detection with a Shifted windows-transformer architecture to reach excellent segmentation results for even the most severely altered glomeruli, arterioles and arteries, even on unseen staining domains from a different nephropathology lab. With accurate automatic segmentation of the decisive renal biopsy compartments in human renal vasculopathies, we have laid the foundation for large-scale compartment-specific machine learning and computer vision analysis of renal biopsy repositories with TMAs.
StyPath: Style-Transfer Data Augmentation For Robust Histology Image Classification
Jul 09, 2020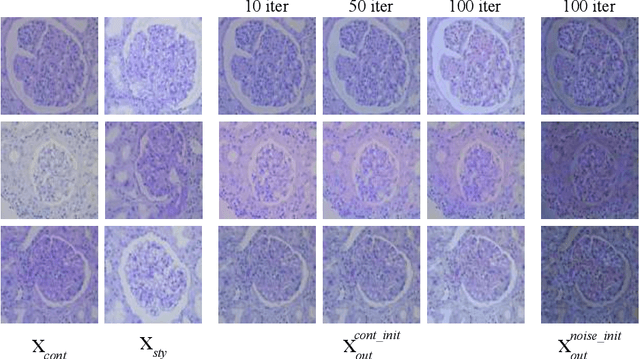
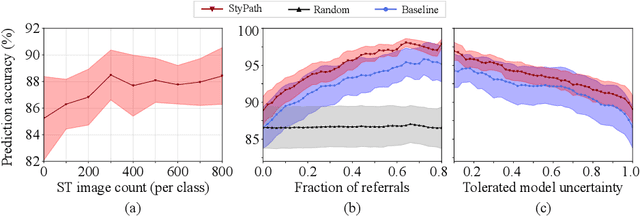
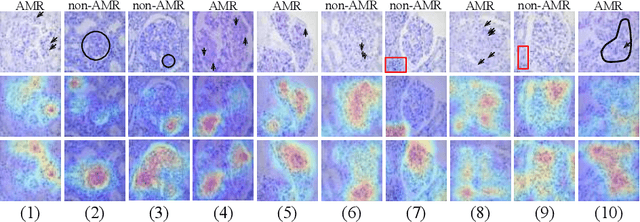
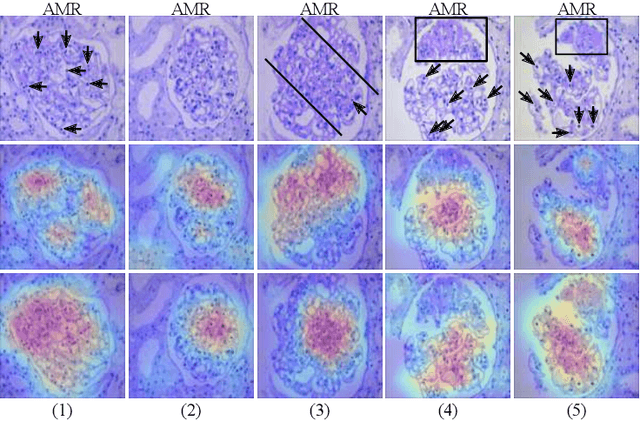
Abstract:The classification of Antibody Mediated Rejection (AMR) in kidney transplant remains challenging even for experienced nephropathologists; this is partly because histological tissue stain analysis is often characterized by low inter-observer agreement and poor reproducibility. One of the implicated causes for inter-observer disagreement is the variability of tissue stain quality between (and within) pathology labs, coupled with the gradual fading of archival sections. Variations in stain colors and intensities can make tissue evaluation difficult for pathologists, ultimately affecting their ability to describe relevant morphological features. Being able to accurately predict the AMR status based on kidney histology images is crucial for improving patient treatment and care. We propose a novel pipeline to build robust deep neural networks for AMR classification based on StyPath, a histological data augmentation technique that leverages a light weight style-transfer algorithm as a means to reduce sample-specific bias. Each image was generated in 1.84 +- 0.03 seconds using a single GTX TITAN V gpu and pytorch, making it faster than other popular histological data augmentation techniques. We evaluated our model using a Monte Carlo (MC) estimate of Bayesian performance and generate an epistemic measure of uncertainty to compare both the baseline and StyPath augmented models. We also generated Grad-CAM representations of the results which were assessed by an experienced nephropathologist; we used this qualitative analysis to elucidate on the assumptions being made by each model. Our results imply that our style-transfer augmentation technique improves histological classification performance (reducing error from 14.8% to 11.5%) and generalization ability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge