Carsten Denkert
Automated Scoring of Nuclear Pleomorphism Spectrum with Pathologist-level Performance in Breast Cancer
Dec 24, 2020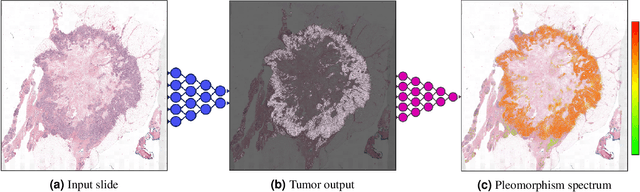
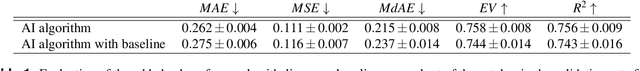
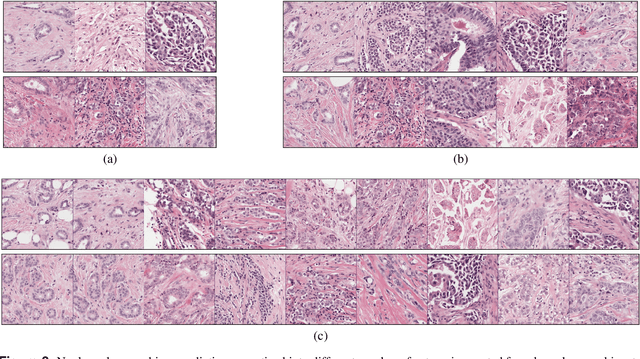
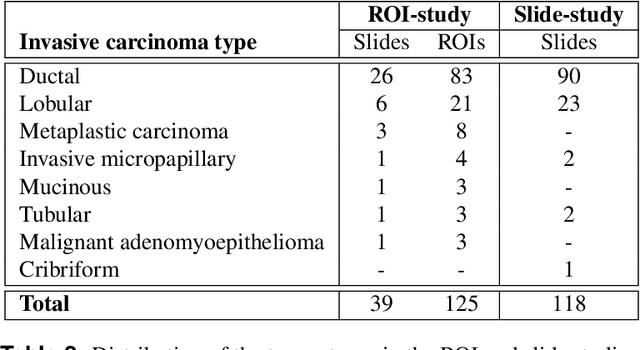
Abstract:Nuclear pleomorphism, defined herein as the extent of abnormalities in the overall appearance of tumor nuclei, is one of the components of the three-tiered breast cancer grading. Given that nuclear pleomorphism reflects a continuous spectrum of variation, we trained a deep neural network on a large variety of tumor regions from the collective knowledge of several pathologists, without constraining the network to the traditional three-category classification. We also motivate an additional approach in which we discuss the additional benefit of normal epithelium as baseline, following the routine clinical practice where pathologists are trained to score nuclear pleomorphism in tumor, having the normal breast epithelium for comparison. In multiple experiments, our fully-automated approach could achieve top pathologist-level performance in select regions of interest as well as at whole slide images, compared to ten and four pathologists, respectively.
Towards computational fluorescence microscopy: Machine learning-based integrated prediction of morphological and molecular tumor profiles
May 28, 2018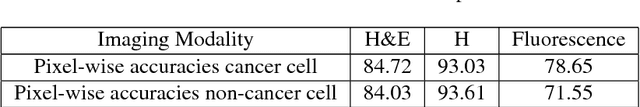
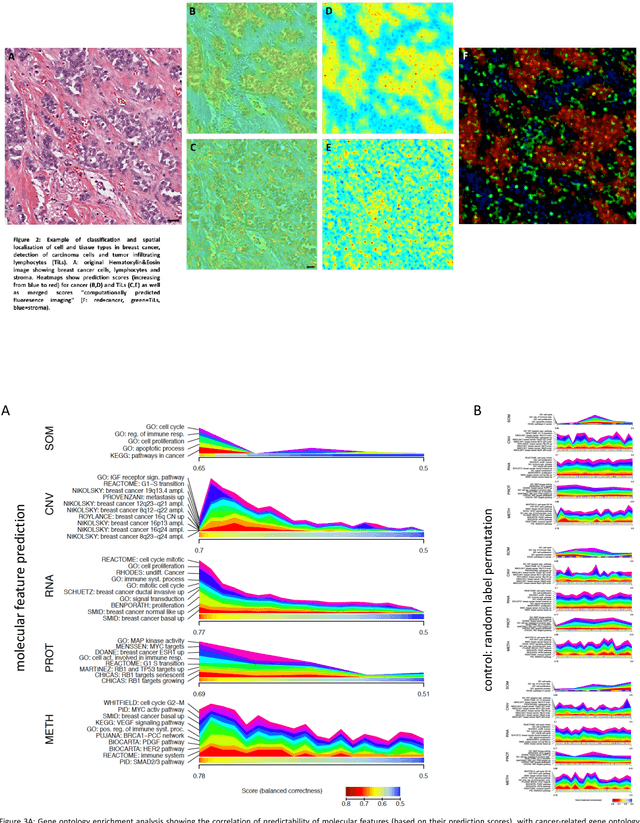
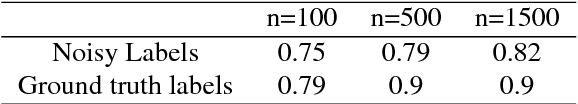

Abstract:Recent advances in cancer research largely rely on new developments in microscopic or molecular profiling techniques offering high level of detail with respect to either spatial or molecular features, but usually not both. Here, we present a novel machine learning-based computational approach that allows for the identification of morphological tissue features and the prediction of molecular properties from breast cancer imaging data. This integration of microanatomic information of tumors with complex molecular profiling data, including protein or gene expression, copy number variation, gene methylation and somatic mutations, provides a novel means to computationally score molecular markers with respect to their relevance to cancer and their spatial associations within the tumor microenvironment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge