Byron C. Wallace
Understanding Clinical Trial Reports: Extracting Medical Entities and Their Relations
Oct 08, 2020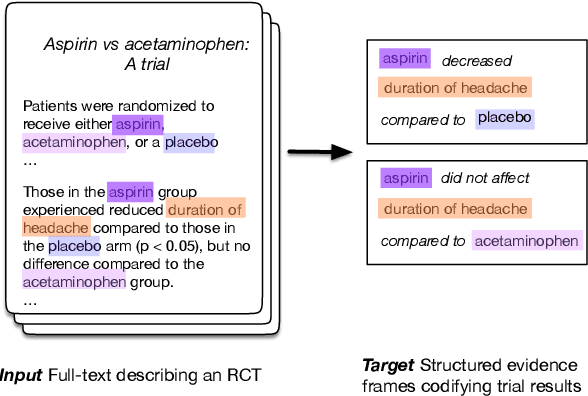
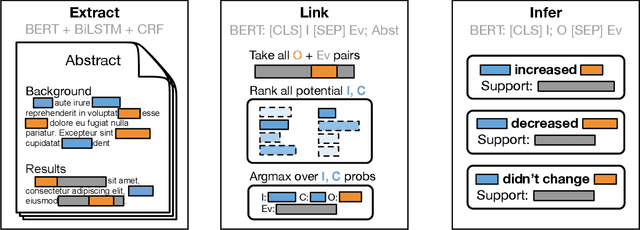

Abstract:The best evidence concerning comparative treatment effectiveness comes from clinical trials, the results of which are reported in unstructured articles. Medical experts must manually extract information from articles to inform decision-making, which is time-consuming and expensive. Here we consider the end-to-end task of both (a) extracting treatments and outcomes from full-text articles describing clinical trials (entity identification) and, (b) inferring the reported results for the former with respect to the latter (relation extraction). We introduce new data for this task, and evaluate models that have recently achieved state-of-the-art results on similar tasks in Natural Language Processing. We then propose a new method motivated by how trial results are typically presented that outperforms these purely data-driven baselines. Finally, we run a fielded evaluation of the model with a non-profit seeking to identify existing drugs that might be re-purposed for cancer, showing the potential utility of end-to-end evidence extraction systems.
Generating (Factual?) Narrative Summaries of RCTs: Experiments with Neural Multi-Document Summarization
Aug 25, 2020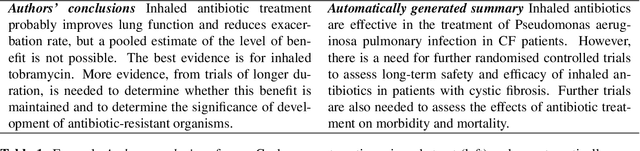
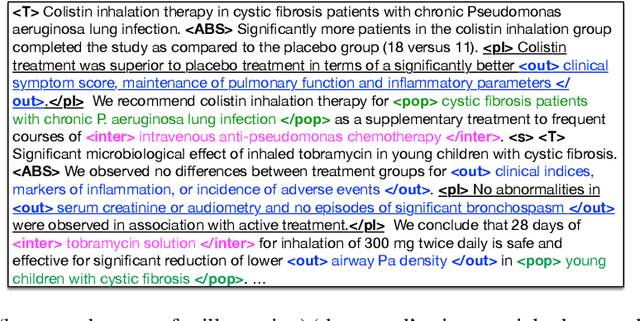

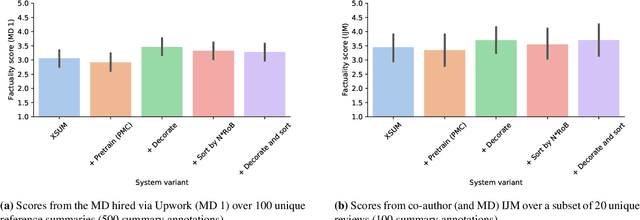
Abstract:We consider the problem of automatically generating a narrative biomedical evidence summary from multiple trial reports. We evaluate modern neural models for abstractive summarization of relevant article abstracts from systematic reviews previously conducted by members of the Cochrane collaboration, using the authors conclusions section of the review abstract as our target. We enlist medical professionals to evaluate generated summaries, and we find that modern summarization systems yield consistently fluent and relevant synopses, but that they are not always factual. We propose new approaches that capitalize on domain-specific models to inform summarization, e.g., by explicitly demarcating snippets of inputs that convey key findings, and emphasizing the reports of large and high-quality trials. We find that these strategies modestly improve the factual accuracy of generated summaries. Finally, we propose a new method for automatically evaluating the factuality of generated narrative evidence syntheses using models that infer the directionality of reported findings.
Semi-Automating Knowledge Base Construction for Cancer Genetics
May 26, 2020
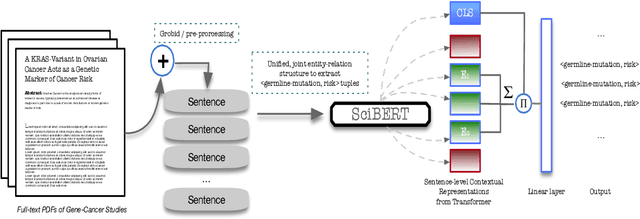
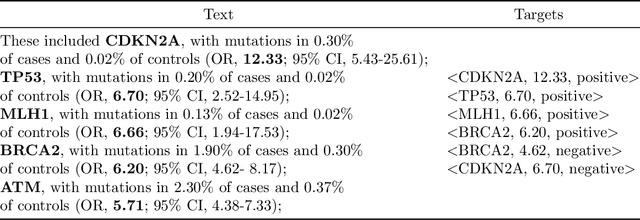

Abstract:In this work, we consider the exponentially growing subarea of genetics in cancer. The need to synthesize and centralize this evidence for dissemination has motivated a team of physicians to manually construct and maintain a knowledge base that distills key results reported in the literature. This is a laborious process that entails reading through full-text articles to understand the study design, assess study quality, and extract the reported cancer risk estimates associated with particular hereditary cancer genes (i.e., penetrance). In this work, we propose models to automatically surface key elements from full-text cancer genetics articles, with the ultimate aim of expediting the manual workflow currently in place. We propose two challenging tasks that are critical for characterizing the findings reported cancer genetics studies: (i) Extracting snippets of text that describe \emph{ascertainment mechanisms}, which in turn inform whether the population studied may introduce bias owing to deviations from the target population; (ii) Extracting reported risk estimates (e.g., odds or hazard ratios) associated with specific germline mutations. The latter task may be viewed as a joint entity tagging and relation extraction problem. To train models for these tasks, we induce distant supervision over tokens and snippets in full-text articles using the manually constructed knowledge base. We propose and evaluate several model variants, including a transformer-based joint entity and relation extraction model to extract <germline mutation, risk-estimate>} pairs. We observe strong empirical performance, highlighting the practical potential for such models to aid KB construction in this space. We ablate components of our model, observing, e.g., that a joint model for <germline mutation, risk-estimate> fares substantially better than a pipelined approach.
Trialstreamer: Mapping and Browsing Medical Evidence in Real-Time
May 21, 2020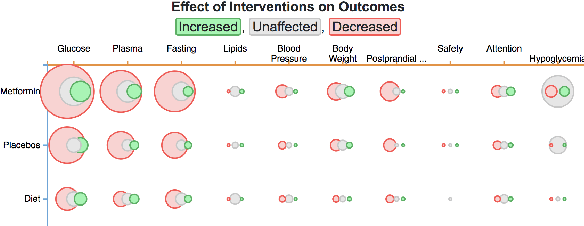
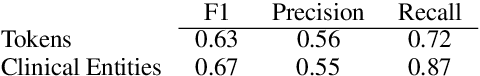
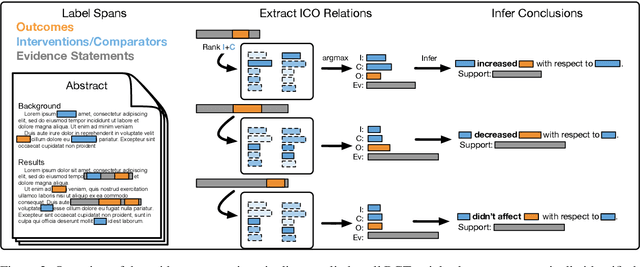
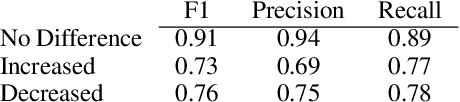
Abstract:We introduce Trialstreamer, a living database of clinical trial reports. Here we mainly describe the evidence extraction component; this extracts from biomedical abstracts key pieces of information that clinicians need when appraising the literature, and also the relations between these. Specifically, the system extracts descriptions of trial participants, the treatments compared in each arm (the interventions), and which outcomes were measured. The system then attempts to infer which interventions were reported to work best by determining their relationship with identified trial outcome measures. In addition to summarizing individual trials, these extracted data elements allow automatic synthesis of results across many trials on the same topic. We apply the system at scale to all reports of randomized controlled trials indexed in MEDLINE, powering the automatic generation of evidence maps, which provide a global view of the efficacy of different interventions combining data from all relevant clinical trials on a topic. We make all code and models freely available alongside a demonstration of the web interface.
Evidence Inference 2.0: More Data, Better Models
May 14, 2020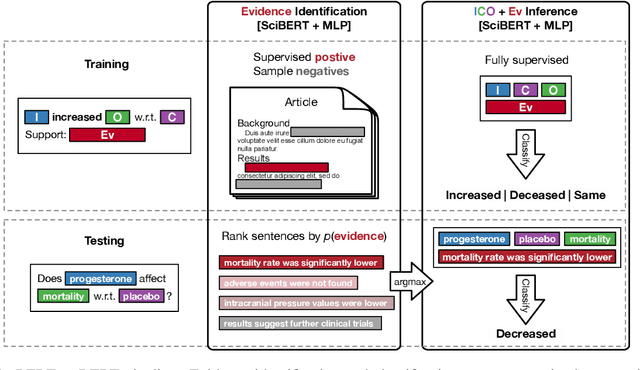
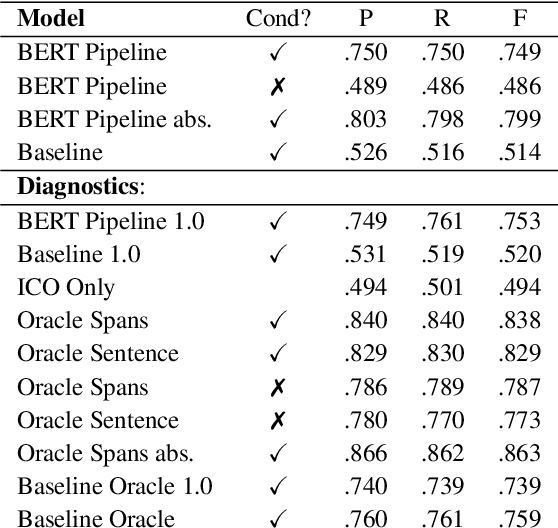
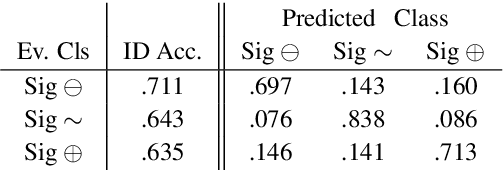
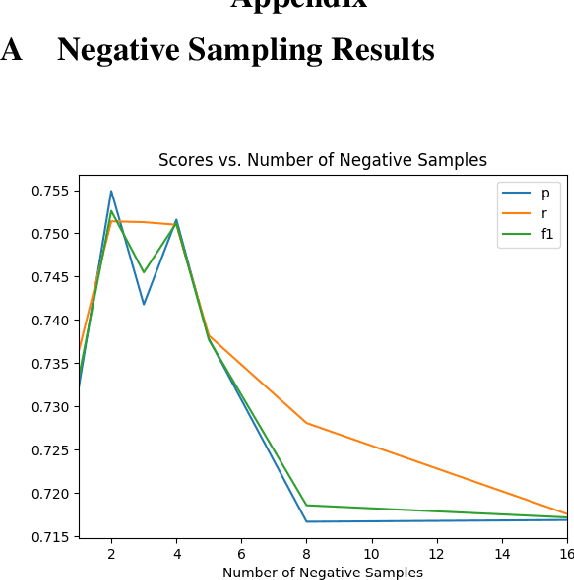
Abstract:How do we most effectively treat a disease or condition? Ideally, we could consult a database of evidence gleaned from clinical trials to answer such questions. Unfortunately, no such database exists; clinical trial results are instead disseminated primarily via lengthy natural language articles. Perusing all such articles would be prohibitively time-consuming for healthcare practitioners; they instead tend to depend on manually compiled systematic reviews of medical literature to inform care. NLP may speed this process up, and eventually facilitate immediate consult of published evidence. The Evidence Inference dataset was recently released to facilitate research toward this end. This task entails inferring the comparative performance of two treatments, with respect to a given outcome, from a particular article (describing a clinical trial) and identifying supporting evidence. For instance: Does this article report that chemotherapy performed better than surgery for five-year survival rates of operable cancers? In this paper, we collect additional annotations to expand the Evidence Inference dataset by 25\%, provide stronger baseline models, systematically inspect the errors that these make, and probe dataset quality. We also release an abstract only (as opposed to full-texts) version of the task for rapid model prototyping. The updated corpus, documentation, and code for new baselines and evaluations are available at http://evidence-inference.ebm-nlp.com/.
Explaining Black Box Predictions and Unveiling Data Artifacts through Influence Functions
May 14, 2020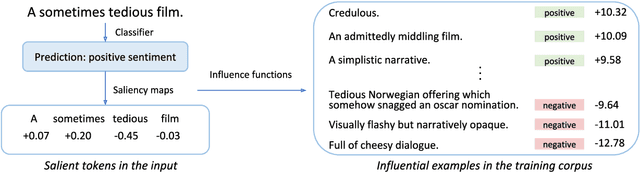


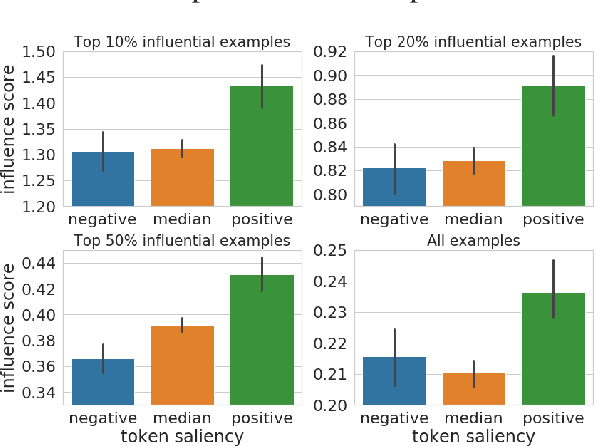
Abstract:Modern deep learning models for NLP are notoriously opaque. This has motivated the development of methods for interpreting such models, e.g., via gradient-based saliency maps or the visualization of attention weights. Such approaches aim to provide explanations for a particular model prediction by highlighting important words in the corresponding input text. While this might be useful for tasks where decisions are explicitly influenced by individual tokens in the input, we suspect that such highlighting is not suitable for tasks where model decisions should be driven by more complex reasoning. In this work, we investigate the use of influence functions for NLP, providing an alternative approach to interpreting neural text classifiers. Influence functions explain the decisions of a model by identifying influential training examples. Despite the promise of this approach, influence functions have not yet been extensively evaluated in the context of NLP, a gap addressed by this work. We conduct a comparison between influence functions and common word-saliency methods on representative tasks. As suspected, we find that influence functions are particularly useful for natural language inference, a task in which 'saliency maps' may not have clear interpretation. Furthermore, we develop a new quantitative measure based on influence functions that can reveal artifacts in training data.
Learning to Faithfully Rationalize by Construction
Apr 30, 2020
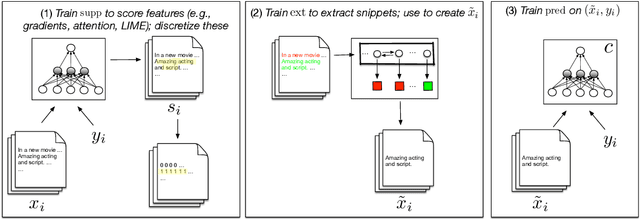
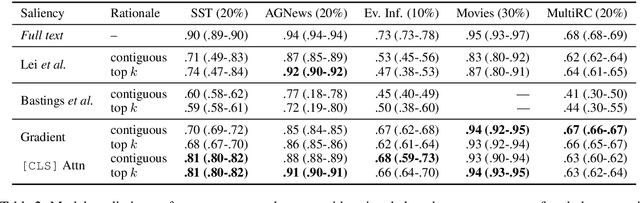
Abstract:In many settings it is important for one to be able to understand why a model made a particular prediction. In NLP this often entails extracting snippets of an input text `responsible for' corresponding model output; when such a snippet comprises tokens that indeed informed the model's prediction, it is a faithful explanation. In some settings, faithfulness may be critical to ensure transparency. Lei et al. (2016) proposed a model to produce faithful rationales for neural text classification by defining independent snippet extraction and prediction modules. However, the discrete selection over input tokens performed by this method complicates training, leading to high variance and requiring careful hyperparameter tuning. We propose a simpler variant of this approach that provides faithful explanations by construction. In our scheme, named FRESH, arbitrary feature importance scores (e.g., gradients from a trained model) are used to induce binary labels over token inputs, which an extractor can be trained to predict. An independent classifier module is then trained exclusively on snippets provided by the extractor; these snippets thus constitute faithful explanations, even if the classifier is arbitrarily complex. In both automatic and manual evaluations we find that variants of this simple framework yield predictive performance superior to `end-to-end' approaches, while being more general and easier to train. Code is available at https://github.com/successar/FRESH
Query-Focused EHR Summarization to Aid Imaging Diagnosis
Apr 26, 2020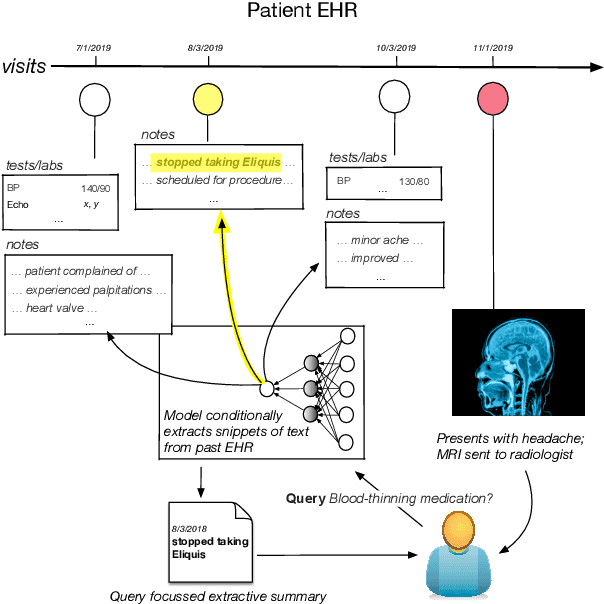
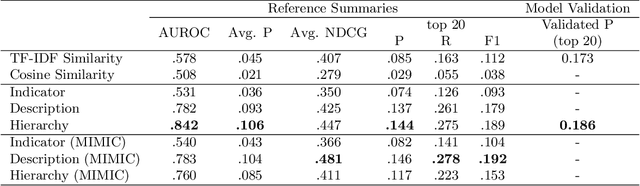
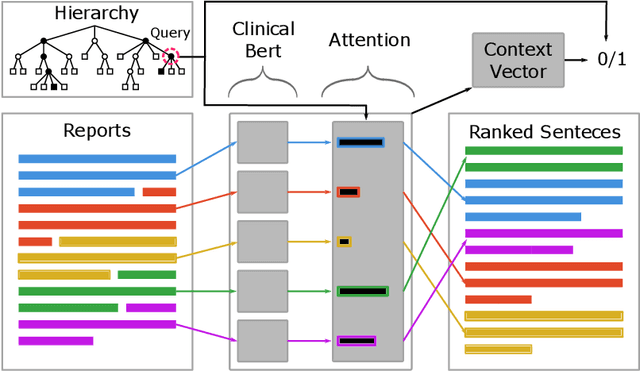
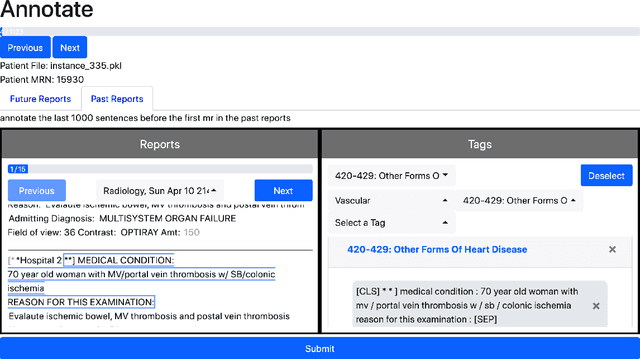
Abstract:Electronic Health Records (EHRs) provide vital contextual information to radiologists and other physicians when making a diagnosis. Unfortunately, because a given patient's record may contain hundreds of notes and reports, identifying relevant information within these in the short time typically allotted to a case is very difficult. We propose and evaluate models that extract relevant text snippets from patient records to provide a rough case summary intended to aid physicians considering one or more diagnoses. This is hard because direct supervision (i.e., physician annotations of snippets relevant to specific diagnoses in medical records) is prohibitively expensive to collect at scale. We propose a distantly supervised strategy in which we use groups of International Classification of Diseases (ICD) codes observed in 'future' records as noisy proxies for 'downstream' diagnoses. Using this we train a transformer-based neural model to perform extractive summarization conditioned on potential diagnoses. This model defines an attention mechanism that is conditioned on potential diagnoses (queries) provided by the diagnosing physician. We train (via distant supervision) and evaluate variants of this model on EHR data from Brigham and Women's Hospital in Boston and MIMIC-III (the latter to facilitate reproducibility). Evaluations performed by radiologists demonstrate that these distantly supervised models yield better extractive summaries than do unsupervised approaches. Such models may aid diagnosis by identifying sentences in past patient reports that are clinically relevant to a potential diagnosis.
Interpretability Analysis for Named Entity Recognition to Understand System Predictions and How They Can Improve
Apr 09, 2020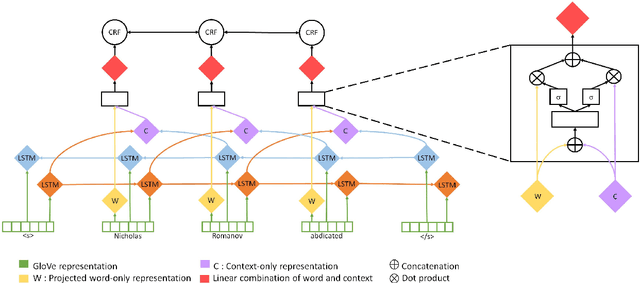
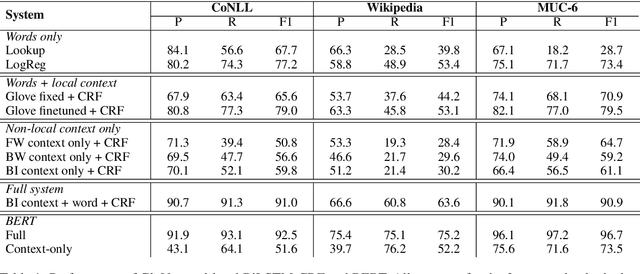
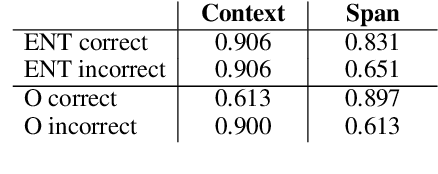

Abstract:Named Entity Recognition systems achieve remarkable performance on domains such as English news. It is natural to ask: What are these models actually learning to achieve this? Are they merely memorizing the names themselves? Or are they capable of interpreting the text and inferring the correct entity type from the linguistic context? We examine these questions by contrasting the performance of several variants of LSTM-CRF architectures for named entity recognition, with some provided only representations of the context as features. We also perform similar experiments for BERT. We find that context representations do contribute to system performance, but that the main factor driving high performance is learning the name tokens themselves. We enlist human annotators to evaluate the feasibility of inferring entity types from the context alone and find that, while people are not able to infer the entity type either for the majority of the errors made by the context-only system, there is some room for improvement. A system should be able to recognize any name in a predictive context correctly and our experiments indicate that current systems may be further improved by such capability.
Entity-Switched Datasets: An Approach to Auditing the In-Domain Robustness of Named Entity Recognition Models
Apr 08, 2020
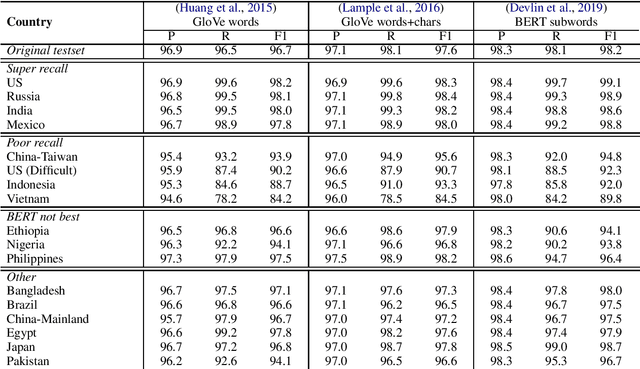
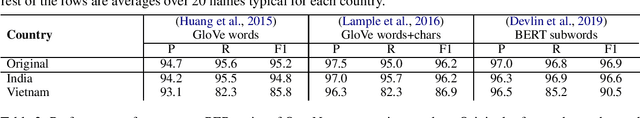
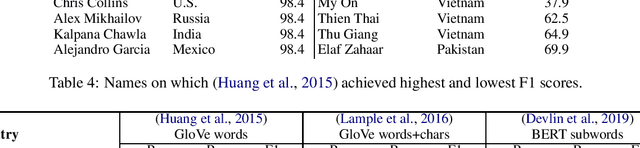
Abstract:Named entity recognition systems perform well on standard datasets comprising English news. But given the paucity of data, it is difficult to draw conclusions about the robustness of systems with respect to recognizing a diverse set of entities. We propose a method for auditing the in-domain robustness of systems, focusing specifically on differences in performance due to the national origin of entities. We create entity-switched datasets, in which named entities in the original texts are replaced by plausible named entities of the same type but of different national origin. We find that state-of-the-art systems' performance vary widely even in-domain: In the same context, entities from certain origins are more reliably recognized than entities from elsewhere. Systems perform best on American and Indian entities, and worst on Vietnamese and Indonesian entities. This auditing approach can facilitate the development of more robust named entity recognition systems, and will allow research in this area to consider fairness criteria that have received heightened attention in other predictive technology work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge