Benjamin Bergner
Digital Health & Machine Learning, Hasso Plattner Institute, University of Potsdam, Germany
Steering Large Reasoning Models towards Concise Reasoning via Flow Matching
Feb 05, 2026Abstract:Large Reasoning Models (LRMs) excel at complex reasoning tasks, but their efficiency is often hampered by overly verbose outputs. Prior steering methods attempt to address this issue by applying a single, global vector to hidden representations -- an approach grounded in the restrictive linear representation hypothesis. In this work, we introduce FlowSteer, a nonlinear steering method that goes beyond uniform linear shifts by learning a complete transformation between the distributions associated with verbose and concise reasoning. This transformation is learned via Flow Matching as a velocity field, enabling precise, input-dependent control over the model's reasoning process. By aligning steered representations with the distribution of concise-reasoning activations, FlowSteer yields more compact reasoning than the linear shifts. Across diverse reasoning benchmarks, FlowSteer demonstrates strong task performance and token efficiency compared to leading inference-time baselines. Our work demonstrates that modeling the full distributional transport with generative techniques offers a more effective and principled foundation for controlling LRMs.
Token Cropr: Faster ViTs for Quite a Few Tasks
Dec 01, 2024



Abstract:The adoption of Vision Transformers (ViTs) in resource-constrained applications necessitates improvements in inference throughput. To this end several token pruning and merging approaches have been proposed that improve efficiency by successively reducing the number of tokens. However, it remains an open problem to design a token reduction method that is fast, maintains high performance, and is applicable to various vision tasks. In this work, we present a token pruner that uses auxiliary prediction heads that learn to select tokens end-to-end based on task relevance. These auxiliary heads can be removed after training, leading to throughput close to that of a random pruner. We evaluate our method on image classification, semantic segmentation, object detection, and instance segmentation, and show speedups of 1.5 to 4x with small drops in performance. As a best case, on the ADE20k semantic segmentation benchmark, we observe a 2x speedup relative to the no-pruning baseline, with a negligible performance penalty of 0.1 median mIoU across 5 seeds.
Think Big, Generate Quick: LLM-to-SLM for Fast Autoregressive Decoding
Feb 26, 2024



Abstract:Large language models (LLMs) have become ubiquitous in practice and are widely used for generation tasks such as translation, summarization and instruction following. However, their enormous size and reliance on autoregressive decoding increase deployment costs and complicate their use in latency-critical applications. In this work, we propose a hybrid approach that combines language models of different sizes to increase the efficiency of autoregressive decoding while maintaining high performance. Our method utilizes a pretrained frozen LLM that encodes all prompt tokens once in parallel, and uses the resulting representations to condition and guide a small language model (SLM), which then generates the response more efficiently. We investigate the combination of encoder-decoder LLMs with both encoder-decoder and decoder-only SLMs from different model families and only require fine-tuning of the SLM. Experiments with various benchmarks show substantial speedups of up to $4\times$, with minor performance penalties of $1-2\%$ for translation and summarization tasks compared to the LLM.
Iterative Patch Selection for High-Resolution Image Recognition
Oct 24, 2022



Abstract:High-resolution images are prevalent in various applications, such as autonomous driving and computer-aided diagnosis. However, training neural networks on such images is computationally challenging and easily leads to out-of-memory errors even on modern GPUs. We propose a simple method, Iterative Patch Selection (IPS), which decouples the memory usage from the input size and thus enables the processing of arbitrarily large images under tight hardware constraints. IPS achieves this by selecting only the most salient patches, which are then aggregated into a global representation for image recognition. For both patch selection and aggregation, a cross-attention based transformer is introduced, which exhibits a close connection to Multiple Instance Learning. Our method demonstrates strong performance and has wide applicability across different domains, training regimes and image sizes while using minimal accelerator memory. For example, we are able to finetune our model on whole-slide images consisting of up to 250k patches (>16 gigapixels) with only 5 GB of GPU VRAM at a batch size of 16.
Less Is More: A Comparison of Active Learning Strategies for 3D Medical Image Segmentation
Jul 02, 2022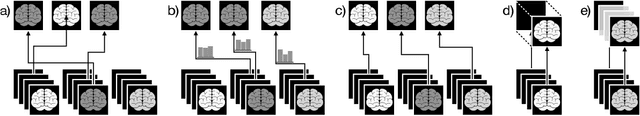
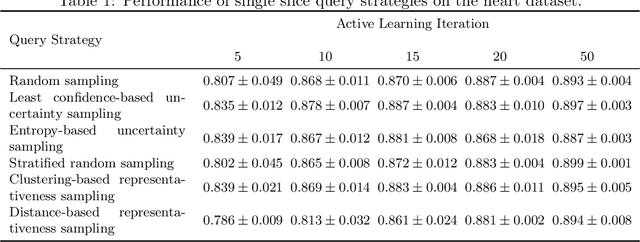
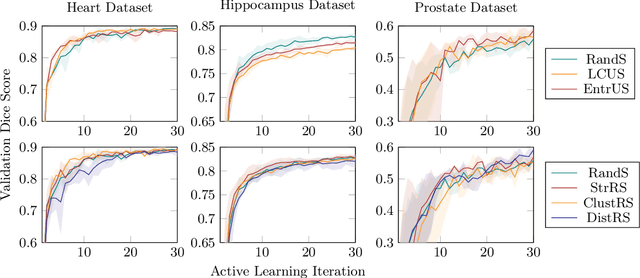
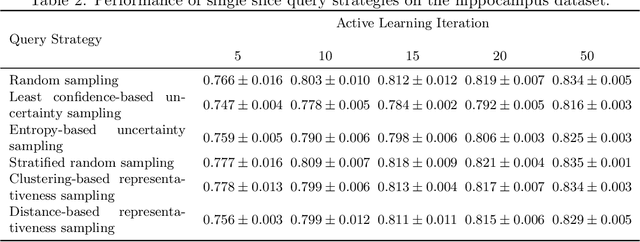
Abstract:Since labeling medical image data is a costly and labor-intensive process, active learning has gained much popularity in the medical image segmentation domain in recent years. A variety of active learning strategies have been proposed in the literature, but their effectiveness is highly dependent on the dataset and training scenario. To facilitate the comparison of existing strategies and provide a baseline for evaluating novel strategies, we evaluate the performance of several well-known active learning strategies on three datasets from the Medical Segmentation Decathlon. Additionally, we consider a strided sampling strategy specifically tailored to 3D image data. We demonstrate that both random and strided sampling act as strong baselines and discuss the advantages and disadvantages of the studied methods. To allow other researchers to compare their work to our results, we provide an open-source framework for benchmarking active learning strategies on a variety of medical segmentation datasets.
Interpretable and Interactive Deep Multiple Instance Learning for Dental Caries Classification in Bitewing X-rays
Dec 17, 2021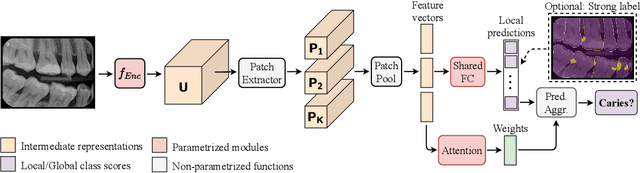

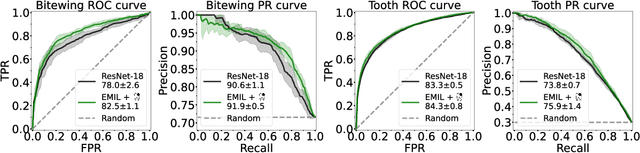
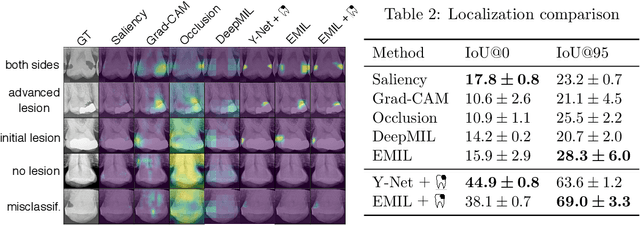
Abstract:We propose a simple and efficient image classification architecture based on deep multiple instance learning, and apply it to the challenging task of caries detection in dental radiographs. Technically, our approach contributes in two ways: First, it outputs a heatmap of local patch classification probabilities despite being trained with weak image-level labels. Second, it is amenable to learning from segmentation labels to guide training. In contrast to existing methods, the human user can faithfully interpret predictions and interact with the model to decide which regions to attend to. Experiments are conducted on a large clinical dataset of $\sim$38k bitewings ($\sim$316k teeth), where we achieve competitive performance compared to various baselines. When guided by an external caries segmentation model, a significant improvement in classification and localization performance is observed.
3D Self-Supervised Methods for Medical Imaging
Jun 06, 2020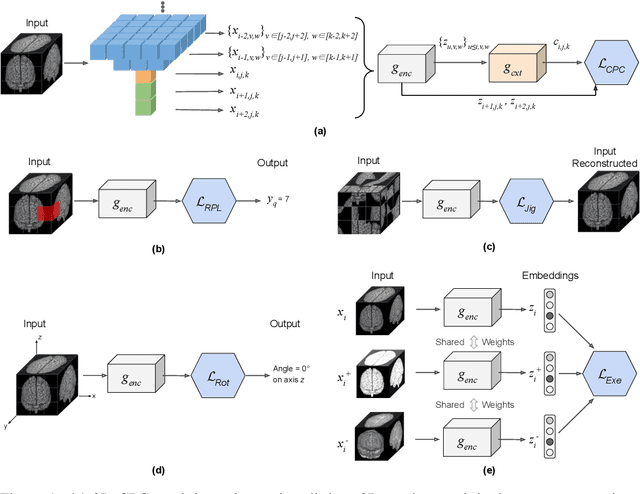
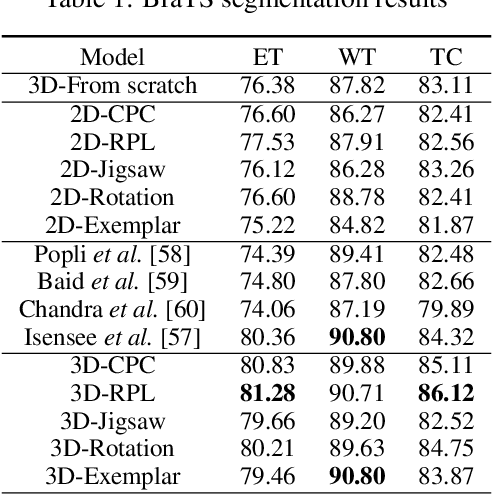
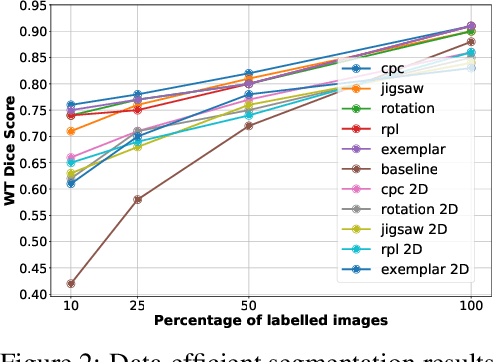
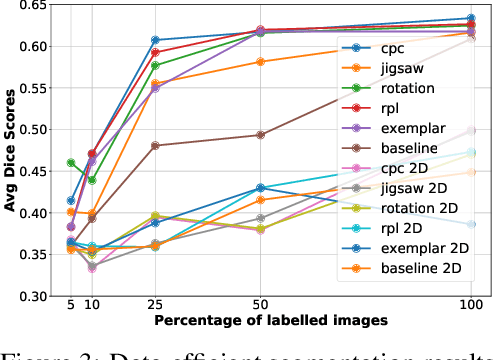
Abstract:Self-supervised learning methods have witnessed a recent surge of interest after proving successful in multiple application fields. In this work, we leverage these techniques, and we propose 3D versions for five different self-supervised methods, in the form of proxy tasks. Our methods facilitate neural network feature learning from unlabeled 3D images, aiming to reduce the required cost for expert annotation. The developed algorithms are 3D Contrastive Predictive Coding, 3D Rotation prediction, 3D Jigsaw puzzles, Relative 3D patch location, and 3D Exemplar networks. Our experiments show that pretraining models with our 3D tasks yields more powerful semantic representations, and enables solving downstream tasks more accurately and efficiently, compared to training the models from scratch and to pretraining them on 2D slices. We demonstrate the effectiveness of our methods on three downstream tasks from the medical imaging domain: i) Brain Tumor Segmentation from 3D MRI, ii) Pancreas Tumor Segmentation from 3D CT, and iii) Diabetic Retinopathy Detection from 2D Fundus images. In each task, we assess the gains in data-efficiency, performance, and speed of convergence. We achieve results competitive to state-of-the-art solutions at a fraction of the computational expense. We also publish the implementations for the 3D and 2D versions of our algorithms as an open-source library, in an effort to allow other researchers to apply and extend our methods on their datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge