Badrinath Roysam
mViSE: A Visual Search Engine for Analyzing Multiplex IHC Brain Tissue Images
Dec 12, 2025Abstract:Whole-slide multiplex imaging of brain tissue generates massive information-dense images that are challenging to analyze and require custom software. We present an alternative query-driven programming-free strategy using a multiplex visual search engine (mViSE) that learns the multifaceted brain tissue chemoarchitecture, cytoarchitecture, and myeloarchitecture. Our divide-and-conquer strategy organizes the data into panels of related molecular markers and uses self-supervised learning to train a multiplex encoder for each panel with explicit visual confirmation of successful learning. Multiple panels can be combined to process visual queries for retrieving similar communities of individual cells or multicellular niches using information-theoretic methods. The retrievals can be used for diverse purposes including tissue exploration, delineating brain regions and cortical cell layers, profiling and comparing brain regions without computer programming. We validated mViSE's ability to retrieve single cells, proximal cell pairs, tissue patches, delineate cortical layers, brain regions and sub-regions. mViSE is provided as an open-source QuPath plug-in.
Weak-to-Strong Generalization Enables Fully Automated De Novo Training of Multi-head Mask-RCNN Model for Segmenting Densely Overlapping Cell Nuclei in Multiplex Whole-slice Brain Images
Dec 12, 2025Abstract:We present a weak to strong generalization methodology for fully automated training of a multi-head extension of the Mask-RCNN method with efficient channel attention for reliable segmentation of overlapping cell nuclei in multiplex cyclic immunofluorescent (IF) whole-slide images (WSI), and present evidence for pseudo-label correction and coverage expansion, the key phenomena underlying weak to strong generalization. This method can learn to segment de novo a new class of images from a new instrument and/or a new imaging protocol without the need for human annotations. We also present metrics for automated self-diagnosis of segmentation quality in production environments, where human visual proofreading of massive WSI images is unaffordable. Our method was benchmarked against five current widely used methods and showed a significant improvement. The code, sample WSI images, and high-resolution segmentation results are provided in open form for community adoption and adaptation.
Few Is Enough: Task-Augmented Active Meta-Learning for Brain Cell Classification
Jul 09, 2020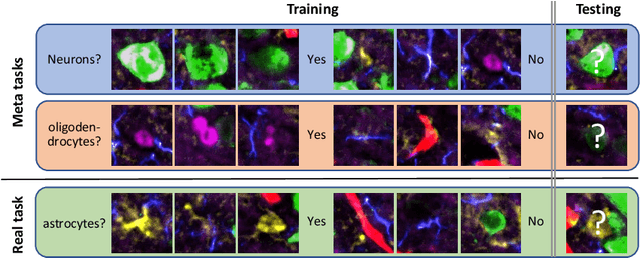
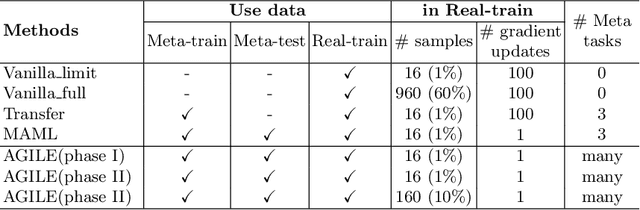
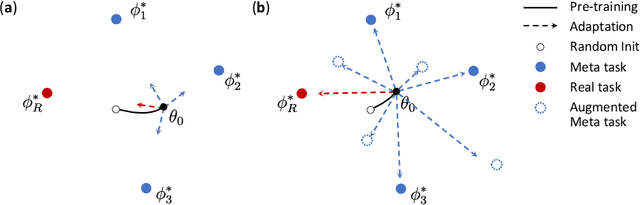
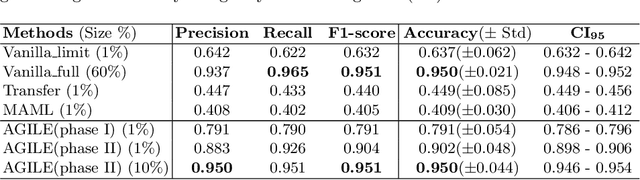
Abstract:Deep Neural Networks (or DNNs) must constantly cope with distribution changes in the input data when the task of interest or the data collection protocol changes. Retraining a network from scratch to combat this issue poses a significant cost. Meta-learning aims to deliver an adaptive model that is sensitive to these underlying distribution changes, but requires many tasks during the meta-training process. In this paper, we propose a tAsk-auGmented actIve meta-LEarning (AGILE) method to efficiently adapt DNNs to new tasks by using a small number of training examples. AGILE combines a meta-learning algorithm with a novel task augmentation technique which we use to generate an initial adaptive model. It then uses Bayesian dropout uncertainty estimates to actively select the most difficult samples when updating the model to a new task. This allows AGILE to learn with fewer tasks and a few informative samples, achieving high performance with a limited dataset. We perform our experiments using the brain cell classification task and compare the results to a plain meta-learning model trained from scratch. We show that the proposed task-augmented meta-learning framework can learn to classify new cell types after a single gradient step with a limited number of training samples. We show that active learning with Bayesian uncertainty can further improve the performance when the number of training samples is extremely small. Using only 1% of the training data and a single update step, we achieved 90% accuracy on the new cell type classification task, a 50% points improvement over a state-of-the-art meta-learning algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge