Antti Rannikko
HistoEncoder: a digital pathology foundation model for prostate cancer
Nov 18, 2024



Abstract:Foundation models are trained on massive amounts of data to distinguish complex patterns and can be adapted to a wide range of downstream tasks with minimal computational resources. Here, we develop a foundation model for prostate cancer digital pathology called HistoEncoder by pre-training on 48 million prostate tissue tile images. We demonstrate that HistoEncoder features extracted from tile images with similar histological patterns map closely together in the feature space. HistoEncoder outperforms models pre-trained with natural images, even without fine-tuning or with 1000 times less training data. We describe two use cases that leverage the capabilities of HistoEncoder by fine-tuning the model with a limited amount of data and computational resources. First, we show how HistoEncoder can be used to automatically annotate large-scale datasets with high accuracy. Second, we combine histomics with commonly used clinical nomograms, significantly improving prostate cancer-specific death survival models. Foundation models such as HistoEncoder can allow organizations with limited resources to build effective clinical software tools without needing extensive datasets or significant amounts of computing.
Exposing and addressing the fragility of neural networks in digital pathology
Jun 30, 2022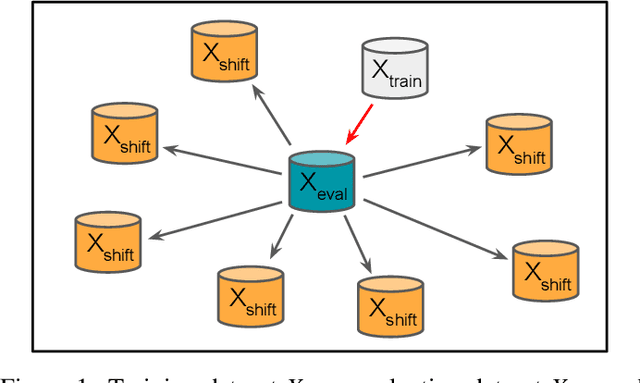
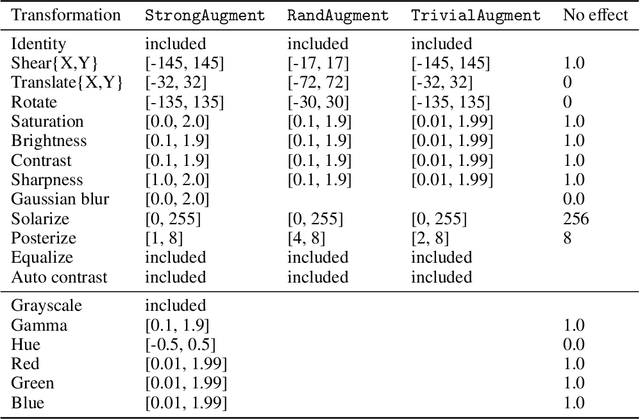
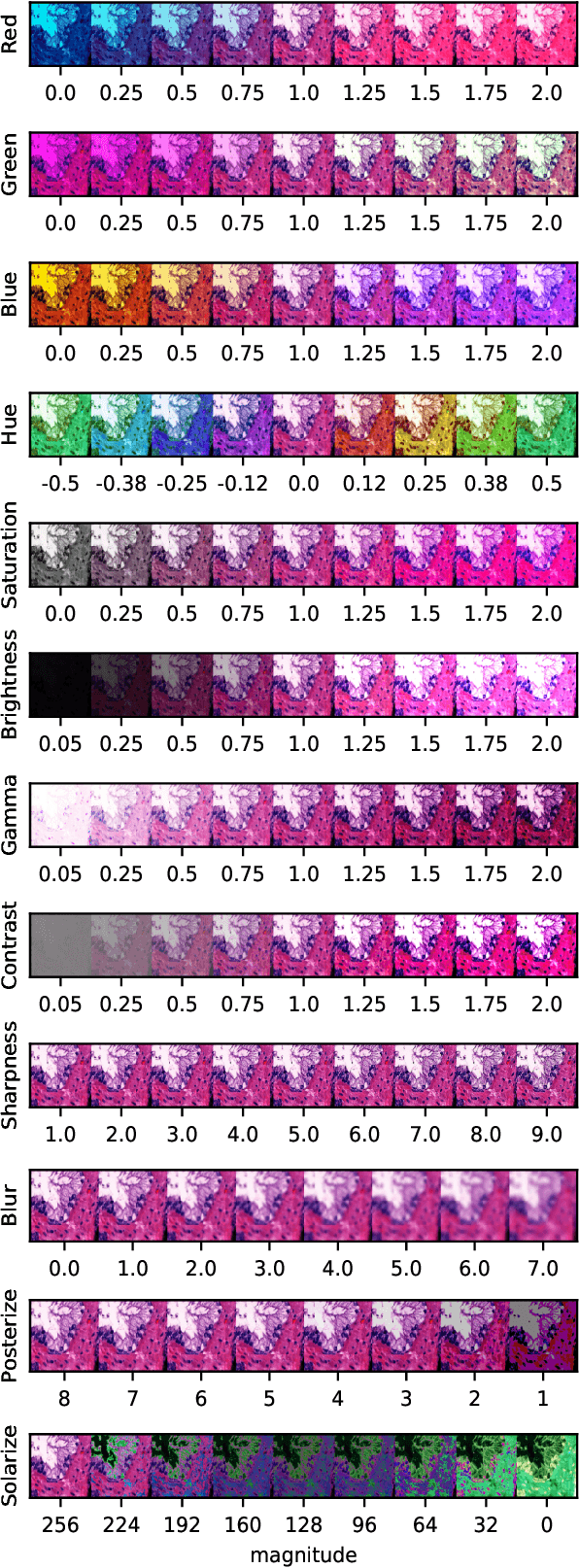
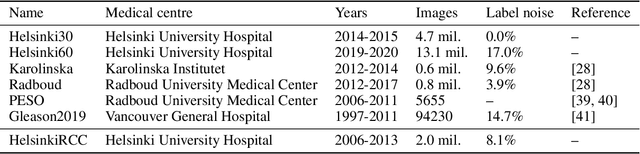
Abstract:Neural networks have achieved impressive results in many medical imaging tasks but often perform substantially worse on out-of-distribution datasets originating from different medical centres or patient cohorts. Evaluating this lack of ability to generalise and address the underlying problem are the two main challenges in developing neural networks intended for clinical practice. In this study, we develop a new method for evaluating neural network models' ability to generalise by generating a large number of distribution-shifted datasets, which can be used to thoroughly investigate their robustness to variability encountered in clinical practice. Compared to external validation, \textit{shifted evaluation} can provide explanations for why neural networks fail on a given dataset, thus offering guidance on how to improve model robustness. With shifted evaluation, we demonstrate that neural networks, trained with state-of-the-art methods, are highly fragile to even small distribution shifts from training data, and in some cases lose all discrimination ability. To address this fragility, we develop an augmentation strategy, explicitly designed to increase neural networks' robustness to distribution shifts. \texttt{StrongAugment} is evaluated with large-scale, heterogeneous histopathology data including five training datasets from two tissue types, 274 distribution-shifted datasets and 20 external datasets from four countries. Neural networks trained with \texttt{StrongAugment} retain similar performance on all datasets, even with distribution shifts where networks trained with current state-of-the-art methods lose all discrimination ability. We recommend using strong augmentation and shifted evaluation to train and evaluate all neural networks intended for clinical practice.
Classification of datasets with imputed missing values: does imputation quality matter?
Jun 16, 2022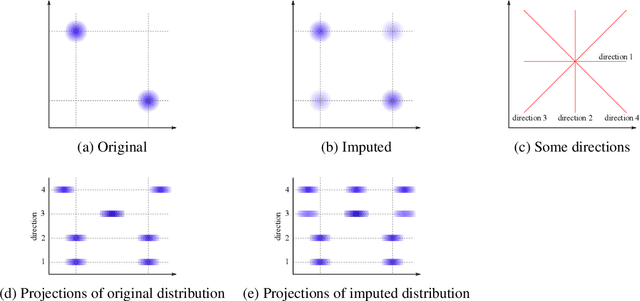
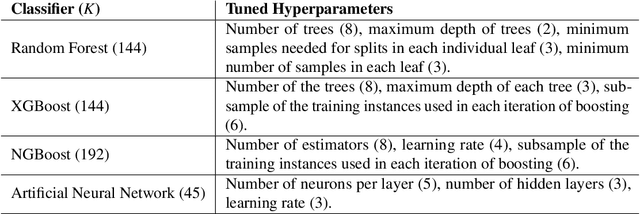

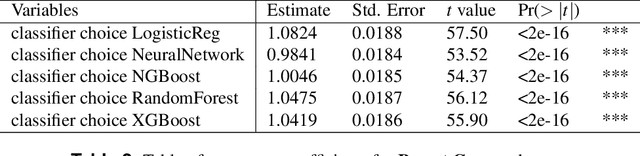
Abstract:Classifying samples in incomplete datasets is a common aim for machine learning practitioners, but is non-trivial. Missing data is found in most real-world datasets and these missing values are typically imputed using established methods, followed by classification of the now complete, imputed, samples. The focus of the machine learning researcher is then to optimise the downstream classification performance. In this study, we highlight that it is imperative to consider the quality of the imputation. We demonstrate how the commonly used measures for assessing quality are flawed and propose a new class of discrepancy scores which focus on how well the method recreates the overall distribution of the data. To conclude, we highlight the compromised interpretability of classifier models trained using poorly imputed data.
Spectral decoupling allows training transferable neural networks in medical imaging
Apr 09, 2021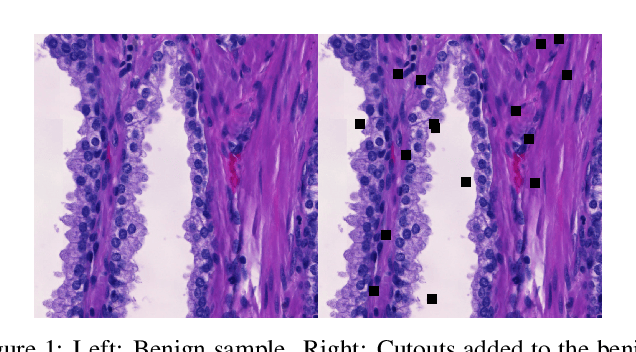

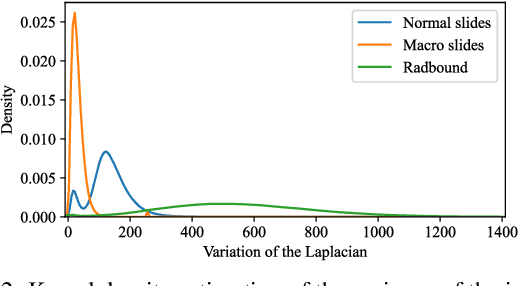
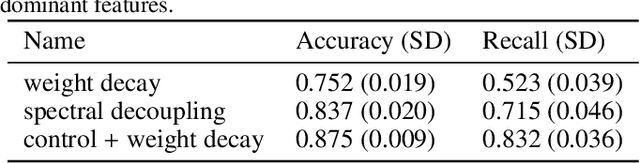
Abstract:Deep neural networks show impressive performance in medical imaging tasks. However, many current networks generalise poorly to data unseen during training, for example data generated by different medical centres. Such behaviour can be caused by networks overfitting easy-to-learn, or statistically dominant, features while disregarding other potentially informative features. Moreover, dominant features can lead to learning spurious correlations. For instance, indistinguishable differences in the sharpness of the images from two different scanners can degrade the performance of the network significantly. To address these challenges, we evaluate the utility of spectral decoupling in the context of medical image classification. Spectral decoupling encourages the neural network to learn more features by simply regularising the networks' unnormalised prediction scores with an L2 penalty. Simulation experiments show that spectral decoupling allows training neural networks on datasets with strong spurious correlations. Networks trained without spectral decoupling do not learn the original task and appear to make false predictions based on the spurious correlations. Spectral decoupling also significantly increases networks' robustness for data distribution shifts. To validate our findings, we train networks with and without spectral decoupling to detect prostate cancer on haematoxylin and eosin stained whole slide images. The networks are then evaluated with data scanned in the same medical centre with two different scanners, and data from a different centre. Networks trained with spectral decoupling increase the accuracy by 10 percentage points over weight decay on the dataset from a different medical centre. Our results show that spectral decoupling allows training robust neural networks to be used across multiple medical centres, and recommend its use in future medical imaging tasks.
Improving Prostate Cancer Detection with Breast Histopathology Images
Mar 14, 2019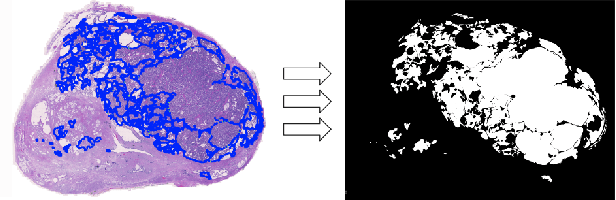
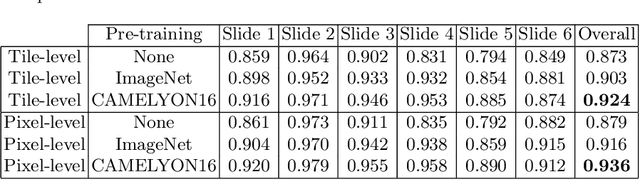
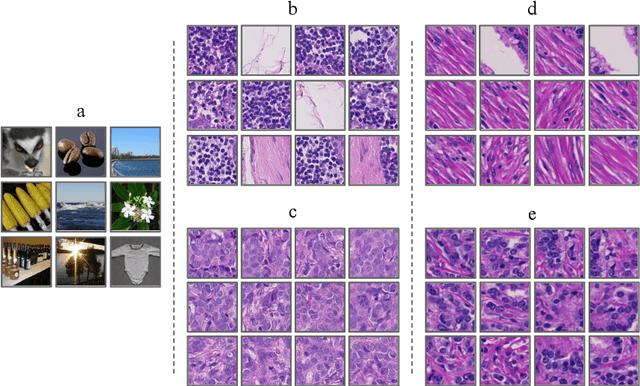
Abstract:Deep neural networks have introduced significant advancements in the field of machine learning-based analysis of digital pathology images including prostate tissue images. With the help of transfer learning, classification and segmentation performance of neural network models have been further increased. However, due to the absence of large, extensively annotated, publicly available prostate histopathology datasets, several previous studies employ datasets from well-studied computer vision tasks such as ImageNet dataset. In this work, we propose a transfer learning scheme from breast histopathology images to improve prostate cancer detection performance. We validate our approach on annotated prostate whole slide images by using a publicly available breast histopathology dataset as pre-training. We show that the proposed cross-cancer approach outperforms transfer learning from ImageNet dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge