Antong Chen
OmniMamba4D: Spatio-temporal Mamba for longitudinal CT lesion segmentation
Apr 13, 2025Abstract:Accurate segmentation of longitudinal CT scans is important for monitoring tumor progression and evaluating treatment responses. However, existing 3D segmentation models solely focus on spatial information. To address this gap, we propose OmniMamba4D, a novel segmentation model designed for 4D medical images (3D images over time). OmniMamba4D utilizes a spatio-temporal tetra-orientated Mamba block to effectively capture both spatial and temporal features. Unlike traditional 3D models, which analyze single-time points, OmniMamba4D processes 4D CT data, providing comprehensive spatio-temporal information on lesion progression. Evaluated on an internal dataset comprising of 3,252 CT scans, OmniMamba4D achieves a competitive Dice score of 0.682, comparable to state-of-the-arts (SOTA) models, while maintaining computational efficiency and better detecting disappeared lesions. This work demonstrates a new framework to leverage spatio-temporal information for longitudinal CT lesion segmentation.
Predicting ulcer in H&E images of inflammatory bowel disease using domain-knowledge-driven graph neural network
Apr 13, 2025Abstract:Inflammatory bowel disease (IBD) involves chronic inflammation of the digestive tract, with treatment options often burdened by adverse effects. Identifying biomarkers for personalized treatment is crucial. While immune cells play a key role in IBD, accurately identifying ulcer regions in whole slide images (WSIs) is essential for characterizing these cells and exploring potential therapeutics. Multiple instance learning (MIL) approaches have advanced WSI analysis but they lack spatial context awareness. In this work, we propose a weakly-supervised model called DomainGCN that employs a graph convolution neural network (GCN) and incorporates domain-specific knowledge of ulcer features, specifically, the presence of epithelium, lymphocytes, and debris for WSI-level ulcer prediction in IBD. We demonstrate that DomainGCN outperforms various state-of-the-art (SOTA) MIL methods and show the added value of domain knowledge.
Multi-dimension unified Swin Transformer for 3D Lesion Segmentation in Multiple Anatomical Locations
Sep 04, 2023Abstract:In oncology research, accurate 3D segmentation of lesions from CT scans is essential for the modeling of lesion growth kinetics. However, following the RECIST criteria, radiologists routinely only delineate each lesion on the axial slice showing the largest transverse area, and delineate a small number of lesions in 3D for research purposes. As a result, we have plenty of unlabeled 3D volumes and labeled 2D images, and scarce labeled 3D volumes, which makes training a deep-learning 3D segmentation model a challenging task. In this work, we propose a novel model, denoted a multi-dimension unified Swin transformer (MDU-ST), for 3D lesion segmentation. The MDU-ST consists of a Shifted-window transformer (Swin-transformer) encoder and a convolutional neural network (CNN) decoder, allowing it to adapt to 2D and 3D inputs and learn the corresponding semantic information in the same encoder. Based on this model, we introduce a three-stage framework: 1) leveraging large amount of unlabeled 3D lesion volumes through self-supervised pretext tasks to learn the underlying pattern of lesion anatomy in the Swin-transformer encoder; 2) fine-tune the Swin-transformer encoder to perform 2D lesion segmentation with 2D RECIST slices to learn slice-level segmentation information; 3) further fine-tune the Swin-transformer encoder to perform 3D lesion segmentation with labeled 3D volumes. The network's performance is evaluated by the Dice similarity coefficient (DSC) and Hausdorff distance (HD) using an internal 3D lesion dataset with 593 lesions extracted from multiple anatomical locations. The proposed MDU-ST demonstrates significant improvement over the competing models. The proposed method can be used to conduct automated 3D lesion segmentation to assist radiomics and tumor growth modeling studies. This paper has been accepted by the IEEE International Symposium on Biomedical Imaging (ISBI) 2023.
Group Equivariant Generative Adversarial Networks
May 04, 2020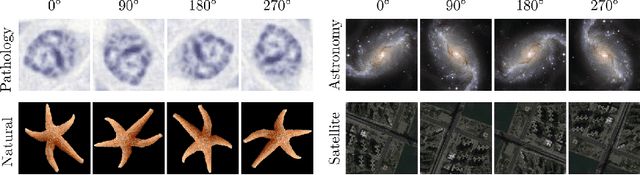
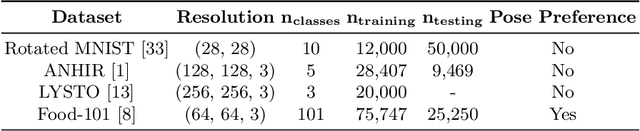
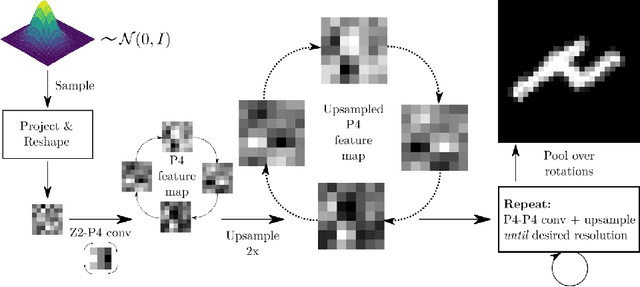
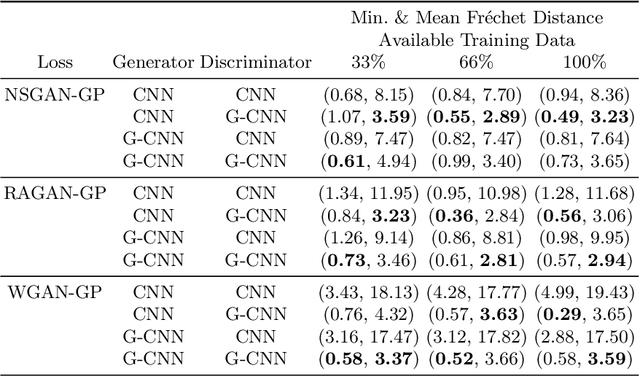
Abstract:Generative adversarial networks are the state of the art for generative modeling in vision, yet are notoriously unstable in practice. This instability is further exacerbated with limited training data. However, in the synthesis of domains such as medical or satellite imaging, it is often overlooked that the image label is invariant to global image symmetries (e.g., rotations and reflections). In this work, we improve gradient feedback between generator and discriminator using an inductive symmetry prior via group-equivariant convolutional networks. We replace convolutional layers with equivalent group-convolutional layers in both generator and discriminator, allowing for better optimization steps and increased expressive power with limited samples. In the process, we extend recent GAN developments to the group-equivariant setting. We demonstrate the utility of our methods by improving both sample fidelity and diversity in the class-conditional synthesis of a diverse set of globally-symmetric imaging modalities.
A deep learning-facilitated radiomics solution for the prediction of lung lesion shrinkage in non-small cell lung cancer trials
Mar 05, 2020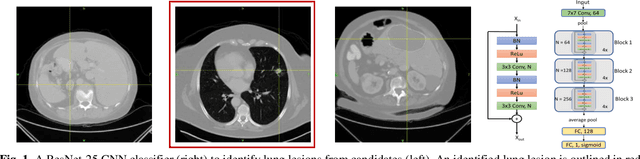
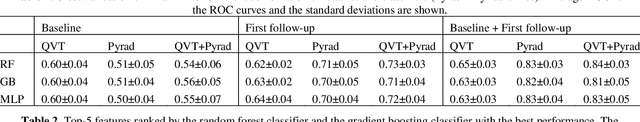
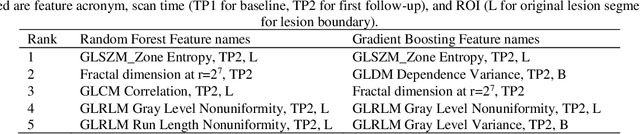

Abstract:Herein we propose a deep learning-based approach for the prediction of lung lesion response based on radiomic features extracted from clinical CT scans of patients in non-small cell lung cancer trials. The approach starts with the classification of lung lesions from the set of primary and metastatic lesions at various anatomic locations. Focusing on the lung lesions, we perform automatic segmentation to extract their 3D volumes. Radiomic features are then extracted from the lesion on the pre-treatment scan and the first follow-up scan to predict which lesions will shrink at least 30% in diameter during treatment (either Pembrolizumab or combinations of chemotherapy and Pembrolizumab), which is defined as a partial response by the Response Evaluation Criteria In Solid Tumors (RECIST) guidelines. A 5-fold cross validation on the training set led to an AUC of 0.84 +/- 0.03, and the prediction on the testing dataset reached AUC of 0.73 +/- 0.02 for the outcome of 30% diameter shrinkage.
Restoration of marker occluded hematoxylin and eosin stained whole slide histology images using generative adversarial networks
Oct 14, 2019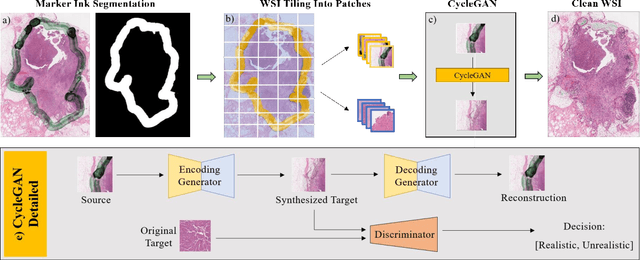
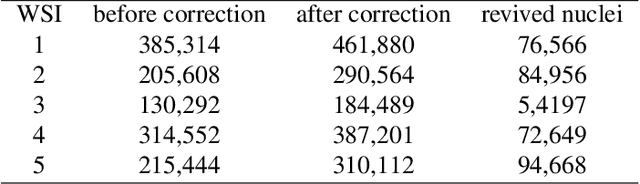
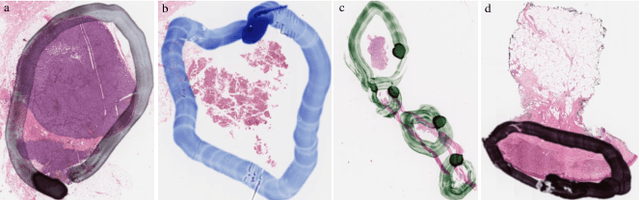
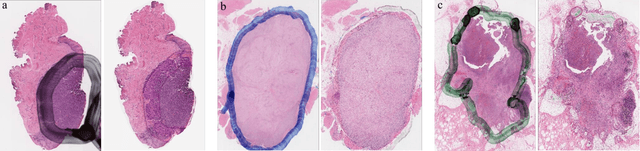
Abstract:It is common for pathologists to annotate specific regions of the tissue, such as tumor, directly on the glass slide with markers. Although this practice was helpful prior to the advent of histology whole slide digitization, it often occludes important details which are increasingly relevant to immuno-oncology due to recent advancements in digital pathology imaging techniques. The current work uses a generative adversarial network with cycle loss to remove these annotations while still maintaining the underlying structure of the tissue by solving an image-to-image translation problem. We train our network on up to 300 whole slide images with marker inks and show that 70% of the corrected image patches are indistinguishable from originally uncontaminated image tissue to a human expert. This portion increases 97% when we replace the human expert with a deep residual network. We demonstrated the fidelity of the method to the original image by calculating the correlation between image gradient magnitudes. We observed a revival of up to 94,000 nuclei per slide in our dataset, the majority of which were located on tissue border.
A multi-level convolutional LSTM model for the segmentation of left ventricle myocardium in infarcted porcine cine MR images
Nov 14, 2018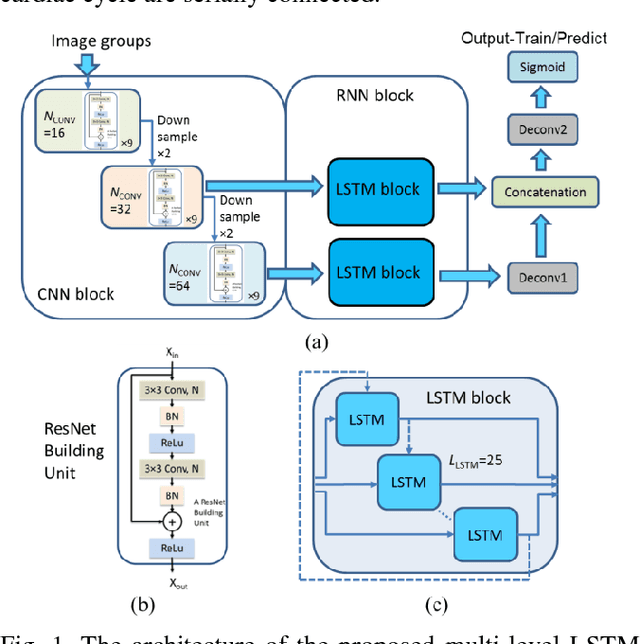
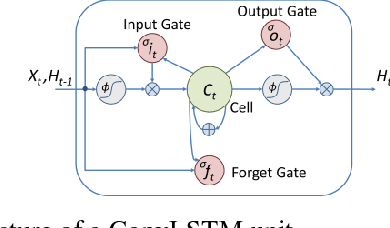
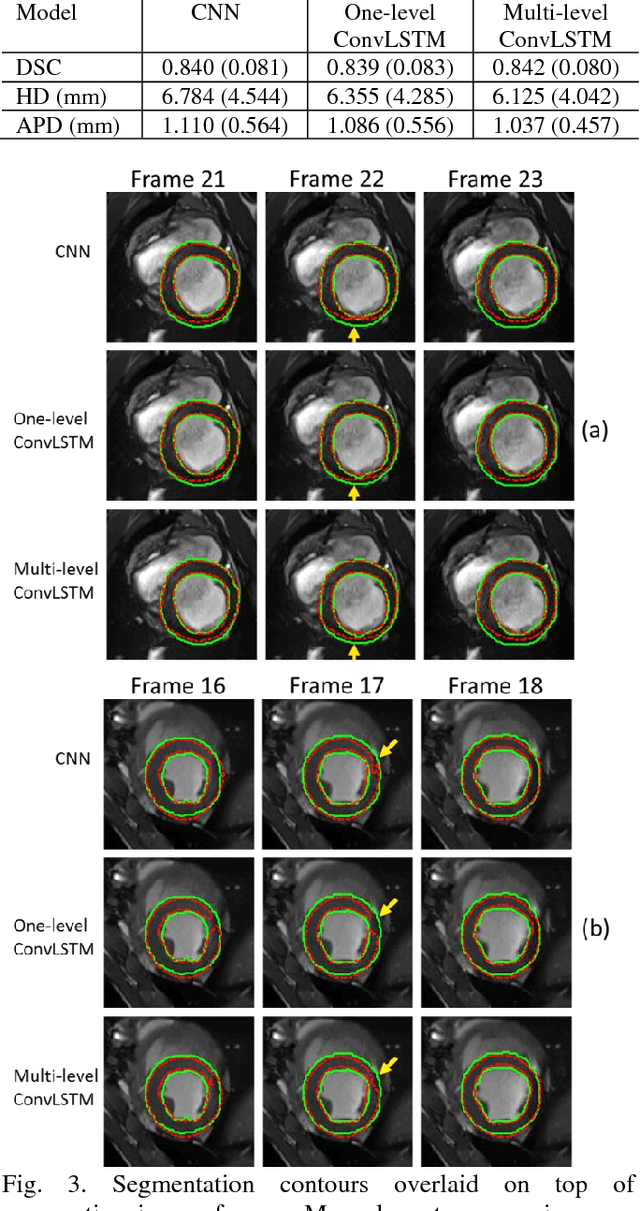
Abstract:Automatic segmentation of left ventricle (LV) myocardium in cardiac short-axis cine MR images acquired on subjects with myocardial infarction is a challenging task, mainly because of the various types of image inhomogeneity caused by the infarctions. Among the approaches proposed to automate the LV myocardium segmentation task, methods based upon deep convolutional neural networks (CNN) have demonstrated their exceptional accuracy and robustness in recent years. However, most of the CNN-based approaches treat the frames in a cardiac cycle independently, which fails to capture the valuable dynamics of heart motion. Herein, an approach based on recurrent neural network (RNN), specifically a multi-level convolutional long short-term memory (ConvLSTM) model, is proposed to take the motion of the heart into consideration. Based on a ResNet-56 CNN, LV-related image features in consecutive frames of a cardiac cycle are extracted at both the low- and high-resolution levels, which are processed by the corresponding multi-level ConvLSTM models to generate the myocardium segmentations. A leave-one-out experiment was carried out on a set of 3,600 cardiac cine MR slices collected in-house for 8 porcine subjects with surgically induced myocardial infarction. Compared with a solely CNN-based approach, the proposed approach demonstrated its superior robustness against image inhomogeneity by incorporating information from adjacent frames. It also outperformed a one-level ConvLSTM approach thanks to its capabilities to take advantage of image features at multiple resolution levels.
A Progressively-trained Scale-invariant and Boundary-aware Deep Neural Network for the Automatic 3D Segmentation of Lung Lesions
Nov 11, 2018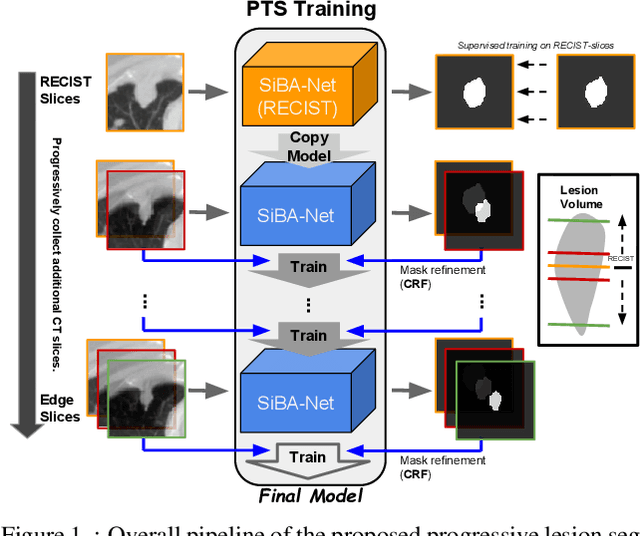
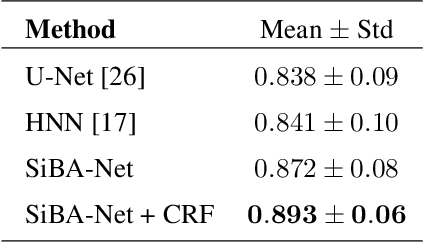
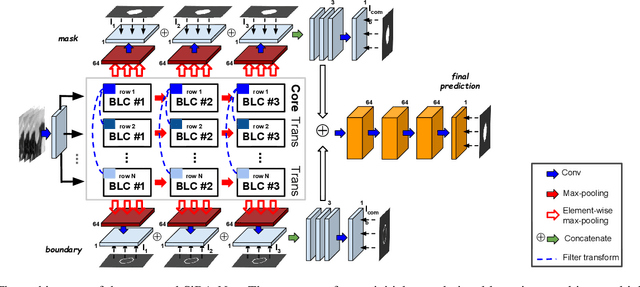
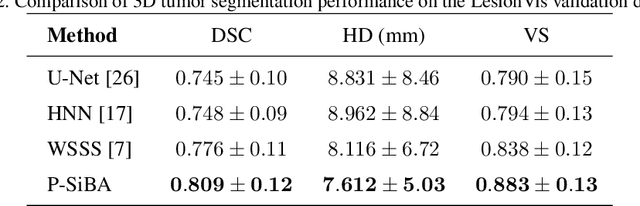
Abstract:Volumetric segmentation of lesions on CT scans is important for many types of analysis, including lesion growth kinetic modeling in clinical trials and machine learning of radiomic features. Manual segmentation is laborious, and impractical for large-scale use. For routine clinical use, and in clinical trials that apply the Response Evaluation Criteria In Solid Tumors (RECIST), clinicians typically outline the boundaries of a lesion on a single slice to extract diameter measurements. In this work, we have collected a large-scale database, named LesionVis, with pixel-wise manual 2D lesion delineations on the RECIST-slices. To extend the 2D segmentations to 3D, we propose a volumetric progressive lesion segmentation (PLS) algorithm to automatically segment the 3D lesion volume from 2D delineations using a scale-invariant and boundary-aware deep convolutional network (SIBA-Net). The SIBA-Net copes with the size transition of a lesion when the PLS progresses from the RECIST-slice to the edge-slices, as well as when performing longitudinal assessment of lesions whose size change over multiple time points. The proposed PLS-SiBA-Net (P-SiBA) approach is assessed on the lung lesion cases from LesionVis. Our experimental results demonstrate that the P-SiBA approach achieves mean Dice similarity coefficients (DSC) of 0.81, which significantly improves 3D segmentation accuracy compared with the approaches proposed previously (highest mean DSC at 0.78 on LesionVis). In summary, by leveraging the limited 2D delineations on the RECIST-slices, P-SiBA is an effective semi-supervised approach to produce accurate lesion segmentations in 3D.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge