Antoine Olivier
IMILIA: interpretable multiple instance learning for inflammation prediction in IBD from H&E whole slide images
Dec 15, 2025Abstract:As the therapeutic target for Inflammatory Bowel Disease (IBD) shifts toward histologic remission, the accurate assessment of microscopic inflammation has become increasingly central for evaluating disease activity and response to treatment. In this work, we introduce IMILIA (Interpretable Multiple Instance Learning for Inflammation Analysis), an end-to-end framework designed for the prediction of inflammation presence in IBD digitized slides stained with hematoxylin and eosin (H&E), followed by the automated computation of markers characterizing tissue regions driving the predictions. IMILIA is composed of an inflammation prediction module, consisting of a Multiple Instance Learning (MIL) model, and an interpretability module, divided in two blocks: HistoPLUS, for cell instance detection, segmentation and classification; and EpiSeg, for epithelium segmentation. IMILIA achieves a cross-validation ROC-AUC of 0.83 on the discovery cohort, and a ROC-AUC of 0.99 and 0.84 on two external validation cohorts. The interpretability module yields biologically consistent insights: tiles with higher predicted scores show increased densities of immune cells (lymphocytes, plasmocytes, neutrophils and eosinophils), whereas lower-scored tiles predominantly contain normal epithelial cells. Notably, these patterns were consistent across all datasets. Code and models to partially replicate the results on the public IBDColEpi dataset can be found at https://github.com/owkin/imilia.
Robust sensitivity control in digital pathology via tile score distribution matching
Feb 28, 2025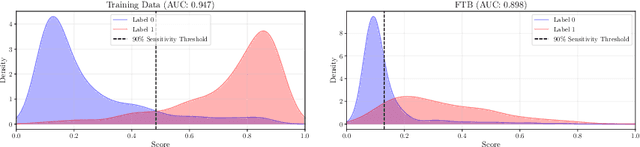
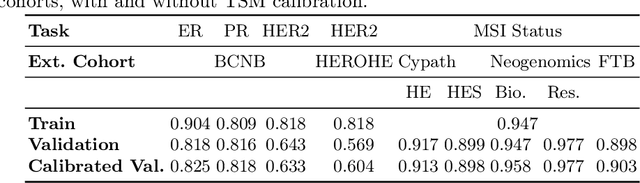
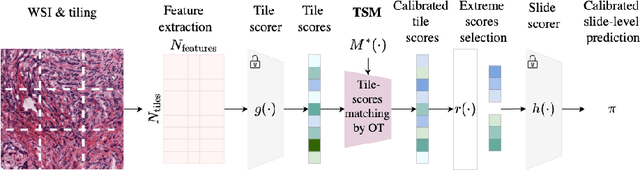
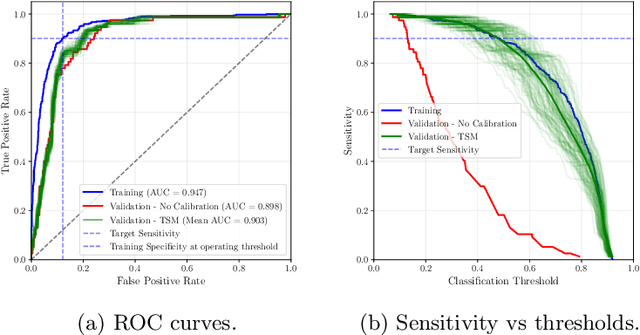
Abstract:Deploying digital pathology models across medical centers is challenging due to distribution shifts. Recent advances in domain generalization improve model transferability in terms of aggregated performance measured by the Area Under Curve (AUC). However, clinical regulations often require to control the transferability of other metrics, such as prescribed sensitivity levels. We introduce a novel approach to control the sensitivity of whole slide image (WSI) classification models, based on optimal transport and Multiple Instance Learning (MIL). Validated across multiple cohorts and tasks, our method enables robust sensitivity control with only a handful of calibration samples, providing a practical solution for reliable deployment of computational pathology systems.
Distilling foundation models for robust and efficient models in digital pathology
Jan 27, 2025Abstract:In recent years, the advent of foundation models (FM) for digital pathology has relied heavily on scaling the pre-training datasets and the model size, yielding large and powerful models. While it resulted in improving the performance on diverse downstream tasks, it also introduced increased computational cost and inference time. In this work, we explore the distillation of a large foundation model into a smaller one, reducing the number of parameters by several orders of magnitude. Leveraging distillation techniques, our distilled model, H0-mini, achieves nearly comparable performance to large FMs at a significantly reduced inference cost. It is evaluated on several public benchmarks, achieving 3rd place on the HEST benchmark and 5th place on the EVA benchmark. Additionally, a robustness analysis conducted on the PLISM dataset demonstrates that our distilled model reaches excellent robustness to variations in staining and scanning conditions, significantly outperforming other state-of-the art models. This opens new perspectives to design lightweight and robust models for digital pathology, without compromising on performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge