Anna Kreshuk
A Generalized Framework for Agglomerative Clustering of Signed Graphs applied to Instance Segmentation
Jun 27, 2019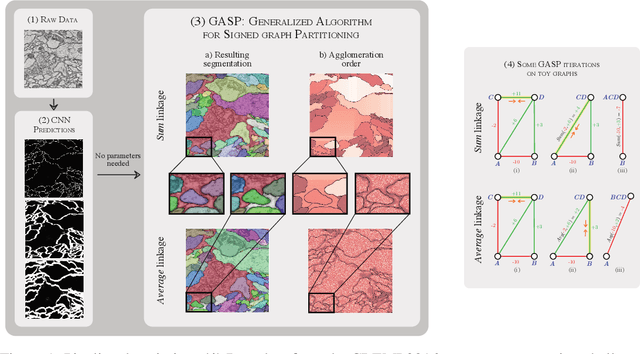
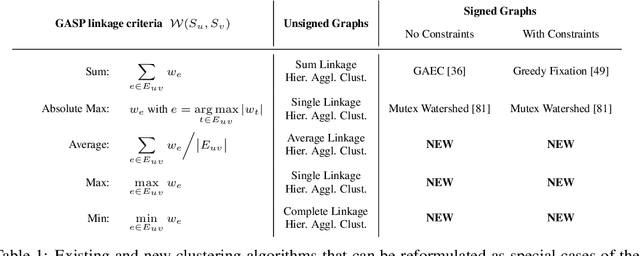
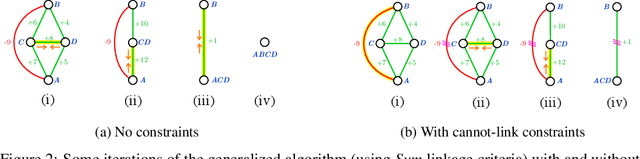
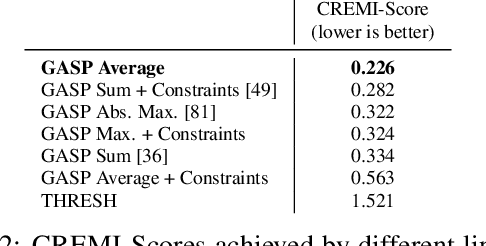
Abstract:We propose a novel theoretical framework that generalizes algorithms for hierarchical agglomerative clustering to weighted graphs with both attractive and repulsive interactions between the nodes. This framework defines GASP, a Generalized Algorithm for Signed graph Partitioning, and allows us to explore many combinations of different linkage criteria and cannot-link constraints. We prove the equivalence of existing clustering methods to some of those combinations, and introduce new algorithms for combinations which have not been studied. An extensive comparison is performed to evaluate properties of the clustering algorithms in the context of instance segmentation in images, including robustness to noise and efficiency. We show how one of the new algorithms proposed in our framework outperforms all previously known agglomerative methods for signed graphs, both on the competitive CREMI 2016 EM segmentation benchmark and on the CityScapes dataset.
Leveraging Domain Knowledge to improve EM image segmentation with Lifted Multicuts
May 25, 2019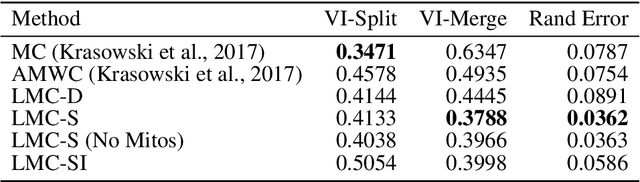
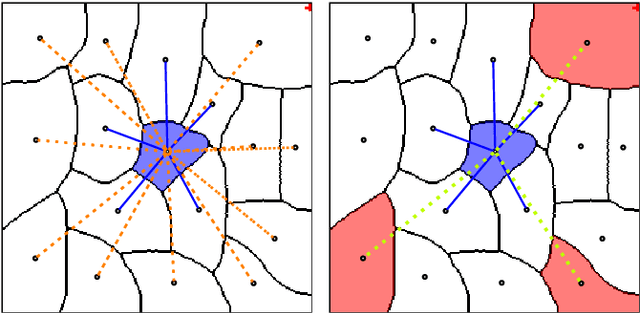
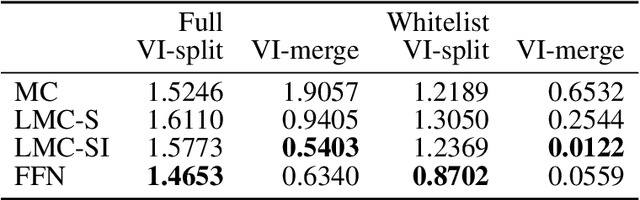
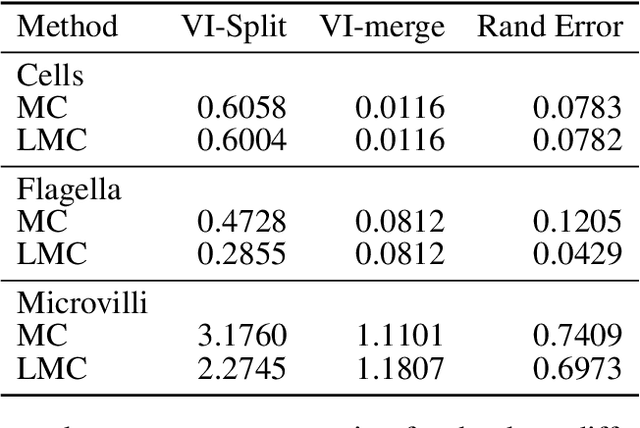
Abstract:The throughput of electron microscopes has increased significantly in recent years, enabling detailed analysis of cell morphology and ultrastructure. Analysis of neural circuits at single-synapse resolution remains the flagship target of this technique, but applications to cell and developmental biology are also starting to emerge at scale. The amount of data acquired in such studies makes manual instance segmentation, a fundamental step in many analysis pipelines, impossible. While automatic segmentation approaches have improved significantly thanks to the adoption of convolutional neural networks, their accuracy still lags behind human annotations and requires additional manual proof-reading. A major hindrance to further improvements is the limited field of view of the segmentation networks preventing them from exploiting the expected cell morphology or other prior biological knowledge which humans use to inform their segmentation decisions. In this contribution, we show how such domain-specific information can be leveraged by expressing it as long-range interactions in a graph partitioning problem known as the lifted multicut problem. Using this formulation, we demonstrate significant improvement in segmentation accuracy for three challenging EM segmentation problems from neuroscience and cell biology.
The Mutex Watershed and its Objective: Efficient, Parameter-Free Image Partitioning
Apr 25, 2019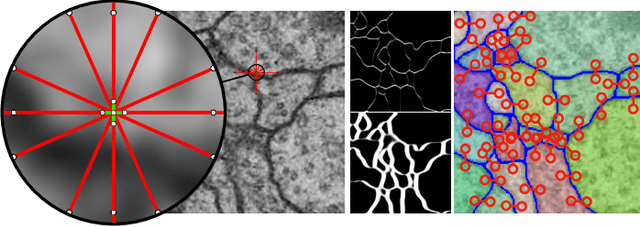
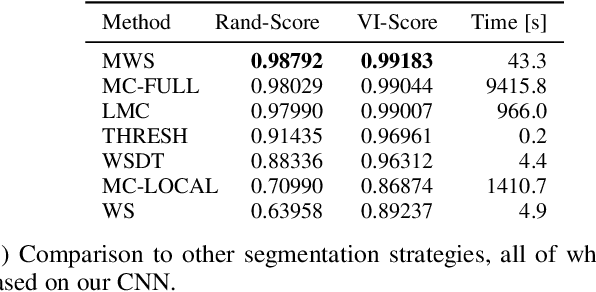
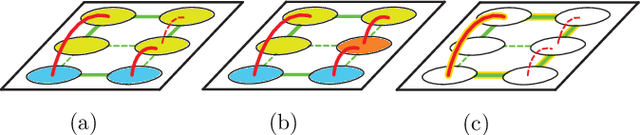
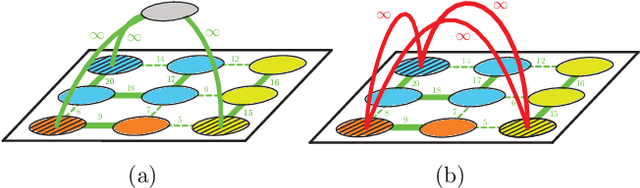
Abstract:Image partitioning, or segmentation without semantics, is the task of decomposing an image into distinct segments, or equivalently to detect closed contours. Most prior work either requires seeds, one per segment; or a threshold; or formulates the task as multicut / correlation clustering, an NP-hard problem. Here, we propose a greedy algorithm for signed graph partitioning, the "Mutex Watershed". Unlike seeded watershed, the algorithm can accommodate not only attractive but also repulsive cues, allowing it to find a previously unspecified number of segments without the need for explicit seeds or a tunable threshold. We also prove that this simple algorithm solves to global optimality an objective function that is intimately related to the multicut / correlation clustering integer linear programming formulation. The algorithm is deterministic, very simple to implement, and has empirically linearithmic complexity. When presented with short-range attractive and long-range repulsive cues from a deep neural network, the Mutex Watershed gives the best results currently known for the competitive ISBI 2012 EM segmentation benchmark.
Domain Adaptive Segmentation in Volume Electron Microscopy Imaging
Oct 23, 2018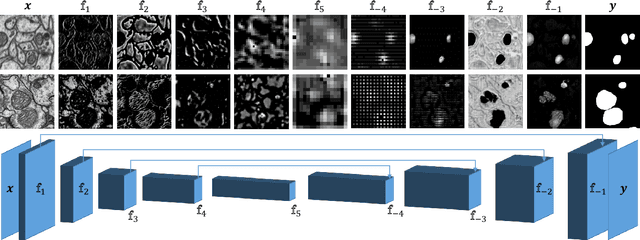
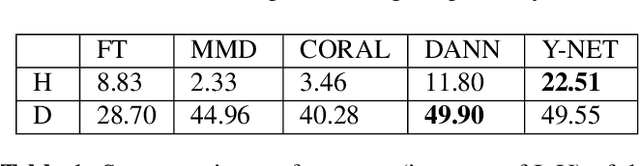
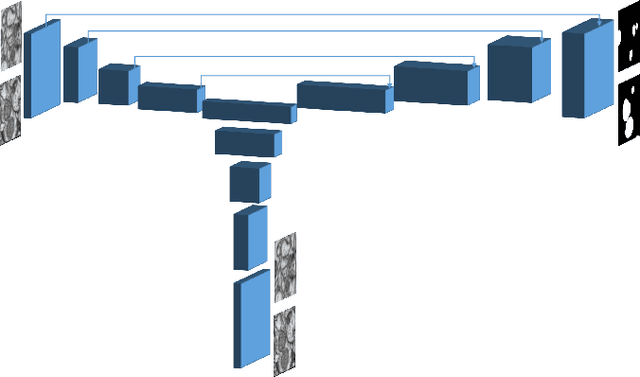
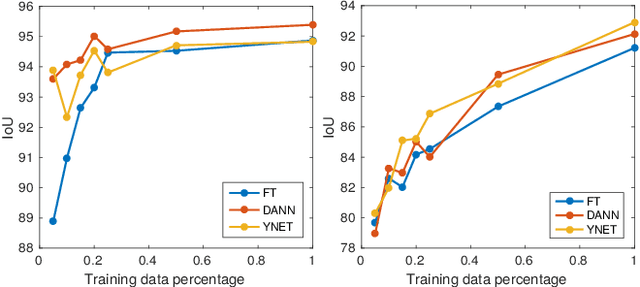
Abstract:In the last years, automated segmentation has become a necessary tool for volume electron microscopy (EM) imaging. So far, the best performing techniques have been largely based on fully supervised encoder-decoder CNNs, requiring a substantial amount of annotated images. Domain Adaptation (DA) aims to alleviate the annotation burden by 'adapting' the networks trained on existing groundtruth data (source domain) to work on a different (target) domain with as little additional annotation as possible. Most DA research is focused on the classification task, whereas volume EM segmentation remains rather unexplored. In this work, we extend recently proposed classification DA techniques to an encoder-decoder layout and propose a novel method that adds a reconstruction decoder to the classical encoder-decoder segmentation in order to align source and target encoder features. The method has been validated on the task of segmenting mitochondria in EM volumes. We have performed DA from brain EM images to HeLa cells and from isotropic FIB/SEM volumes to anisotropic TEM volumes. In all cases, the proposed method has outperformed the extended classification DA techniques and the finetuning baseline. An implementation of our work can be found on https://github.com/JorisRoels/domain-adaptive-segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge