Anders Nymark Christensen
Weight Space Correlation Analysis: Quantifying Feature Utilization in Deep Learning Models
Dec 15, 2025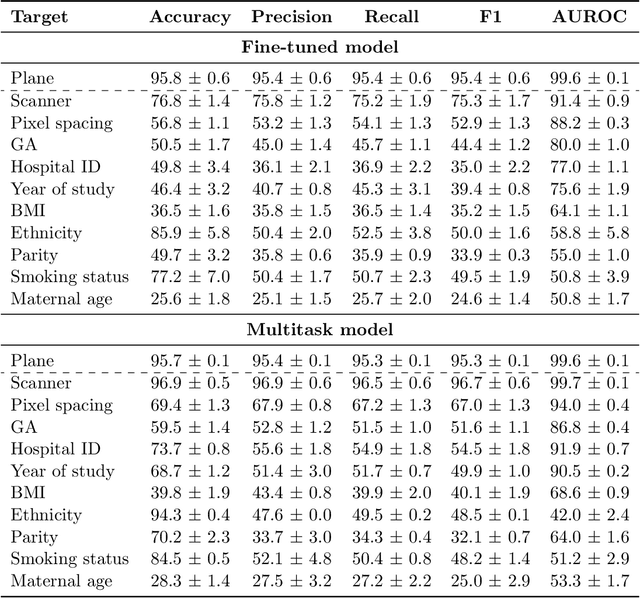
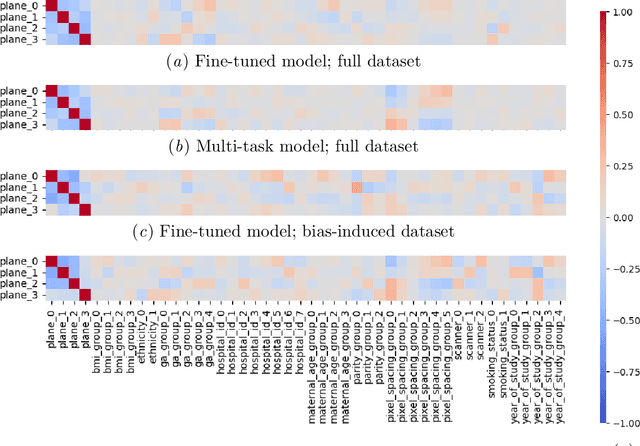
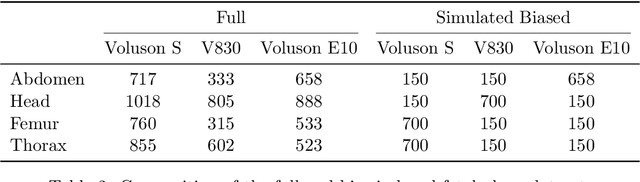

Abstract:Deep learning models in medical imaging are susceptible to shortcut learning, relying on confounding metadata (e.g., scanner model) that is often encoded in image embeddings. The crucial question is whether the model actively utilizes this encoded information for its final prediction. We introduce Weight Space Correlation Analysis, an interpretable methodology that quantifies feature utilization by measuring the alignment between the classification heads of a primary clinical task and auxiliary metadata tasks. We first validate our method by successfully detecting artificially induced shortcut learning. We then apply it to probe the feature utilization of an SA-SonoNet model trained for Spontaneous Preterm Birth (sPTB) prediction. Our analysis confirmed that while the embeddings contain substantial metadata, the sPTB classifier's weight vectors were highly correlated with clinically relevant factors (e.g., birth weight) but decoupled from clinically irrelevant acquisition factors (e.g. scanner). Our methodology provides a tool to verify model trustworthiness, demonstrating that, in the absence of induced bias, the clinical model selectively utilizes features related to the genuine clinical signal.
General Methods Make Great Domain-specific Foundation Models: A Case-study on Fetal Ultrasound
Jun 24, 2025Abstract:With access to large-scale, unlabeled medical datasets, researchers are confronted with two questions: Should they attempt to pretrain a custom foundation model on this medical data, or use transfer-learning from an existing generalist model? And, if a custom model is pretrained, are novel methods required? In this paper we explore these questions by conducting a case-study, in which we train a foundation model on a large regional fetal ultrasound dataset of 2M images. By selecting the well-established DINOv2 method for pretraining, we achieve state-of-the-art results on three fetal ultrasound datasets, covering data from different countries, classification, segmentation, and few-shot tasks. We compare against a series of models pretrained on natural images, ultrasound images, and supervised baselines. Our results demonstrate two key insights: (i) Pretraining on custom data is worth it, even if smaller models are trained on less data, as scaling in natural image pretraining does not translate to ultrasound performance. (ii) Well-tuned methods from computer vision are making it feasible to train custom foundation models for a given medical domain, requiring no hyperparameter tuning and little methodological adaptation. Given these findings, we argue that a bias towards methodological innovation should be avoided when developing domain specific foundation models under common computational resource constraints.
Determining Fetal Orientations From Blind Sweep Ultrasound Video
Apr 09, 2025Abstract:Cognitive demands of fetal ultrasound examinations pose unique challenges among clinicians. With the goal of providing an assistive tool, we developed an automated pipeline for predicting fetal orientation from ultrasound videos acquired following a simple blind sweep protocol. Leveraging on a pre-trained head detection and segmentation model, this is achieved by first determining the fetal presentation (cephalic or breech) with a template matching approach, followed by the fetal lie (facing left or right) by analyzing the spatial distribution of segmented brain anatomies. Evaluation on a dataset of third-trimester ultrasound scans demonstrated the promising accuracy of our pipeline. This work distinguishes itself by introducing automated fetal lie prediction and by proposing an assistive paradigm that augments sonographer expertise rather than replacing it. Future research will focus on enhancing acquisition efficiency, and exploring real-time clinical integration to improve workflow and support for obstetric clinicians.
Fast Sphericity and Roundness approximation in 2D and 3D using Local Thickness
Apr 08, 2025Abstract:Sphericity and roundness are fundamental measures used for assessing object uniformity in 2D and 3D images. However, using their strict definition makes computation costly. As both 2D and 3D microscopy imaging datasets grow larger, there is an increased demand for efficient algorithms that can quantify multiple objects in large volumes. We propose a novel approach for extracting sphericity and roundness based on the output of a local thickness algorithm. For sphericity, we simplify the surface area computation by modeling objects as spheroids/ellipses of varying lengths and widths of mean local thickness. For roundness, we avoid a complex corner curvature determination process by approximating it with local thickness values on the contour/surface of the object. The resulting methods provide an accurate representation of the exact measures while being significantly faster than their existing implementations.
MozzaVID: Mozzarella Volumetric Image Dataset
Dec 06, 2024Abstract:Influenced by the complexity of volumetric imaging, there is a shortage of established datasets useful for benchmarking volumetric deep-learning models. As a consequence, new and existing models are not easily comparable, limiting the development of architectures optimized specifically for volumetric data. To counteract this trend, we introduce MozzaVID - a large, clean, and versatile volumetric classification dataset. Our dataset contains X-ray computed tomography (CT) images of mozzarella microstructure and enables the classification of 25 cheese types and 149 cheese samples. We provide data in three different resolutions, resulting in three dataset instances containing from 591 to 37,824 images. While being general-purpose, the dataset also facilitates investigating mozzarella structure properties. The structure of food directly affects its functional properties and thus its consumption experience. Understanding food structure helps tune the production and mimicking it enables sustainable alternatives to animal-derived food products. The complex and disordered nature of food structures brings a unique challenge, where a choice of appropriate imaging method, scale, and sample size is not trivial. With this dataset we aim to address these complexities, contributing to more robust structural analysis models. The dataset can be downloaded from: https://archive.compute.dtu.dk/files/public/projects/MozzaVID/.
Unsupervised Detection of Fetal Brain Anomalies using Denoising Diffusion Models
Aug 07, 2024Abstract:Congenital malformations of the brain are among the most common fetal abnormalities that impact fetal development. Previous anomaly detection methods on ultrasound images are based on supervised learning, rely on manual annotations, and risk missing underrepresented categories. In this work, we frame fetal brain anomaly detection as an unsupervised task using diffusion models. To this end, we employ an inpainting-based Noise Agnostic Anomaly Detection approach that identifies the abnormality using diffusion-reconstructed fetal brain images from multiple noise levels. Our approach only requires normal fetal brain ultrasound images for training, addressing the limited availability of abnormal data. Our experiments on a real-world clinical dataset show the potential of using unsupervised methods for fetal brain anomaly detection. Additionally, we comprehensively evaluate how different noise types affect diffusion models in the fetal anomaly detection domain.
Deployment of Deep Learning Model in Real World Clinical Setting: A Case Study in Obstetric Ultrasound
Mar 22, 2024

Abstract:Despite the rapid development of AI models in medical image analysis, their validation in real-world clinical settings remains limited. To address this, we introduce a generic framework designed for deploying image-based AI models in such settings. Using this framework, we deployed a trained model for fetal ultrasound standard plane detection, and evaluated it in real-time sessions with both novice and expert users. Feedback from these sessions revealed that while the model offers potential benefits to medical practitioners, the need for navigational guidance was identified as a key area for improvement. These findings underscore the importance of early deployment of AI models in real-world settings, leading to insights that can guide the refinement of the model and system based on actual user feedback.
Diffusion-based Iterative Counterfactual Explanations for Fetal Ultrasound Image Quality Assessment
Mar 13, 2024



Abstract:Obstetric ultrasound image quality is crucial for accurate diagnosis and monitoring of fetal health. However, producing high-quality standard planes is difficult, influenced by the sonographer's expertise and factors like the maternal BMI or the fetus dynamics. In this work, we propose using diffusion-based counterfactual explainable AI to generate realistic high-quality standard planes from low-quality non-standard ones. Through quantitative and qualitative evaluation, we demonstrate the effectiveness of our method in producing plausible counterfactuals of increased quality. This shows future promise both for enhancing training of clinicians by providing visual feedback, as well as for improving image quality and, consequently, downstream diagnosis and monitoring.
Shortcut Learning in Medical Image Segmentation
Mar 11, 2024Abstract:Shortcut learning is a phenomenon where machine learning models prioritize learning simple, potentially misleading cues from data that do not generalize well beyond the training set. While existing research primarily investigates this in the realm of image classification, this study extends the exploration of shortcut learning into medical image segmentation. We demonstrate that clinical annotations such as calipers, and the combination of zero-padded convolutions and center-cropped training sets in the dataset can inadvertently serve as shortcuts, impacting segmentation accuracy. We identify and evaluate the shortcut learning on two different but common medical image segmentation tasks. In addition, we suggest strategies to mitigate the influence of shortcut learning and improve the generalizability of the segmentation models. By uncovering the presence and implications of shortcuts in medical image segmentation, we provide insights and methodologies for evaluating and overcoming this pervasive challenge and call for attention in the community for shortcuts in segmentation.
Learning semantic image quality for fetal ultrasound from noisy ranking annotation
Feb 13, 2024



Abstract:We introduce the notion of semantic image quality for applications where image quality relies on semantic requirements. Working in fetal ultrasound, where ranking is challenging and annotations are noisy, we design a robust coarse-to-fine model that ranks images based on their semantic image quality and endow our predicted rankings with an uncertainty estimate. To annotate rankings on training data, we design an efficient ranking annotation scheme based on the merge sort algorithm. Finally, we compare our ranking algorithm to a number of state-of-the-art ranking algorithms on a challenging fetal ultrasound quality assessment task, showing the superior performance of our method on the majority of rank correlation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge