Amith Khandakar
CASR-Net: An Image Processing-focused Deep Learning-based Coronary Artery Segmentation and Refinement Network for X-ray Coronary Angiogram
Oct 31, 2025Abstract:Early detection of coronary artery disease (CAD) is critical for reducing mortality and improving patient treatment planning. While angiographic image analysis from X-rays is a common and cost-effective method for identifying cardiac abnormalities, including stenotic coronary arteries, poor image quality can significantly impede clinical diagnosis. We present the Coronary Artery Segmentation and Refinement Network (CASR-Net), a three-stage pipeline comprising image preprocessing, segmentation, and refinement. A novel multichannel preprocessing strategy combining CLAHE and an improved Ben Graham method provides incremental gains, increasing Dice Score Coefficient (DSC) by 0.31-0.89% and Intersection over Union (IoU) by 0.40-1.16% compared with using the techniques individually. The core innovation is a segmentation network built on a UNet with a DenseNet121 encoder and a Self-organized Operational Neural Network (Self-ONN) based decoder, which preserves the continuity of narrow and stenotic vessel branches. A final contour refinement module further suppresses false positives. Evaluated with 5-fold cross-validation on a combination of two public datasets that contain both healthy and stenotic arteries, CASR-Net outperformed several state-of-the-art models, achieving an IoU of 61.43%, a DSC of 76.10%, and clDice of 79.36%. These results highlight a robust approach to automated coronary artery segmentation, offering a valuable tool to support clinicians in diagnosis and treatment planning.
Harnessing Smartphone Sensors for Enhanced Road Safety: A Comprehensive Dataset and Review
Nov 13, 2024Abstract:Severe collisions can result from aggressive driving and poor road conditions, emphasizing the need for effective monitoring to ensure safety. Smartphones, with their array of built-in sensors, offer a practical and affordable solution for road-sensing. However, the lack of reliable, standardized datasets has hindered progress in assessing road conditions and driving patterns. This study addresses this gap by introducing a comprehensive dataset derived from smartphone sensors, which surpasses existing datasets by incorporating a diverse range of sensors including accelerometer, gyroscope, magnetometer, GPS, gravity, orientation, and uncalibrated sensors. These sensors capture extensive parameters such as acceleration force, gravitation, rotation rate, magnetic field strength, and vehicle speed, providing a detailed understanding of road conditions and driving behaviors. The dataset is designed to enhance road safety, infrastructure maintenance, traffic management, and urban planning. By making this dataset available to the community, the study aims to foster collaboration, inspire further research, and facilitate the development of innovative solutions in intelligent transportation systems.
Novel Interpretable and Robust Web-based AI Platform for Phishing Email Detection
May 19, 2024



Abstract:Phishing emails continue to pose a significant threat, causing financial losses and security breaches. This study addresses limitations in existing research, such as reliance on proprietary datasets and lack of real-world application, by proposing a high-performance machine learning model for email classification. Utilizing a comprehensive and largest available public dataset, the model achieves a f1 score of 0.99 and is designed for deployment within relevant applications. Additionally, Explainable AI (XAI) is integrated to enhance user trust. This research offers a practical and highly accurate solution, contributing to the fight against phishing by empowering users with a real-time web-based application for phishing email detection.
BIO-CXRNET: A Robust Multimodal Stacking Machine Learning Technique for Mortality Risk Prediction of COVID-19 Patients using Chest X-Ray Images and Clinical Data
Jun 15, 2022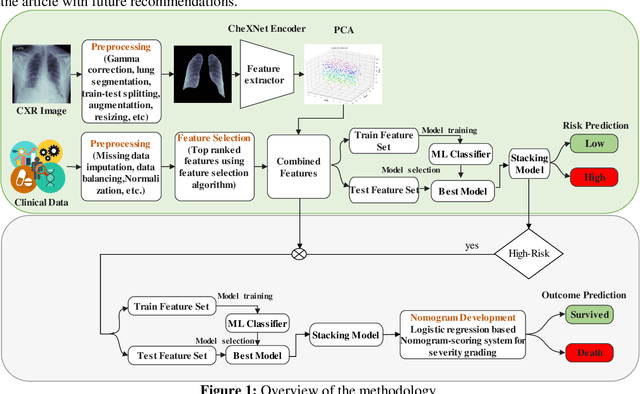
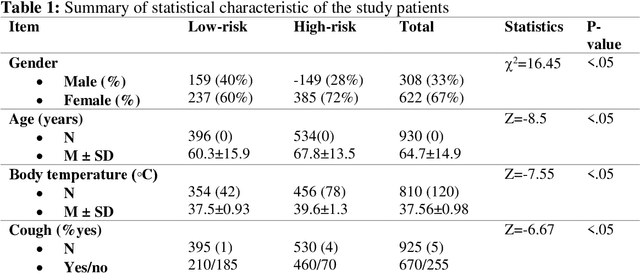
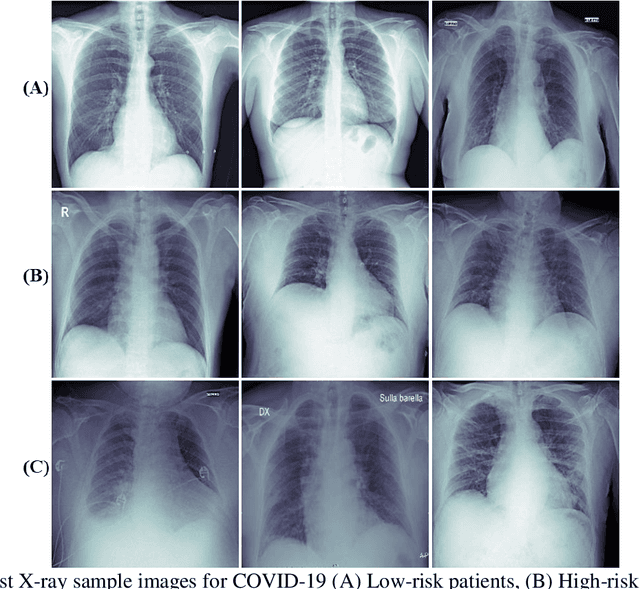
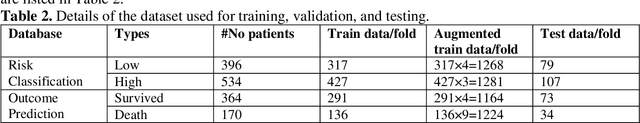
Abstract:Fast and accurate detection of the disease can significantly help in reducing the strain on the healthcare facility of any country to reduce the mortality during any pandemic. The goal of this work is to create a multimodal system using a novel machine learning framework that uses both Chest X-ray (CXR) images and clinical data to predict severity in COVID-19 patients. In addition, the study presents a nomogram-based scoring technique for predicting the likelihood of death in high-risk patients. This study uses 25 biomarkers and CXR images in predicting the risk in 930 COVID-19 patients admitted during the first wave of COVID-19 (March-June 2020) in Italy. The proposed multimodal stacking technique produced the precision, sensitivity, and F1-score, of 89.03%, 90.44%, and 89.03%, respectively to identify low or high-risk patients. This multimodal approach improved the accuracy by 6% in comparison to the CXR image or clinical data alone. Finally, nomogram scoring system using multivariate logistic regression -- was used to stratify the mortality risk among the high-risk patients identified in the first stage. Lactate Dehydrogenase (LDH), O2 percentage, White Blood Cells (WBC) Count, Age, and C-reactive protein (CRP) were identified as useful predictor using random forest feature selection model. Five predictors parameters and a CXR image based nomogram score was developed for quantifying the probability of death and categorizing them into two risk groups: survived (<50%), and death (>=50%), respectively. The multi-modal technique was able to predict the death probability of high-risk patients with an F1 score of 92.88 %. The area under the curves for the development and validation cohorts are 0.981 and 0.939, respectively.
Motion Artifacts Correction from Single-Channel EEG and fNIRS Signals using Novel Wavelet Packet Decomposition in Combination with Canonical Correlation Analysis
Apr 09, 2022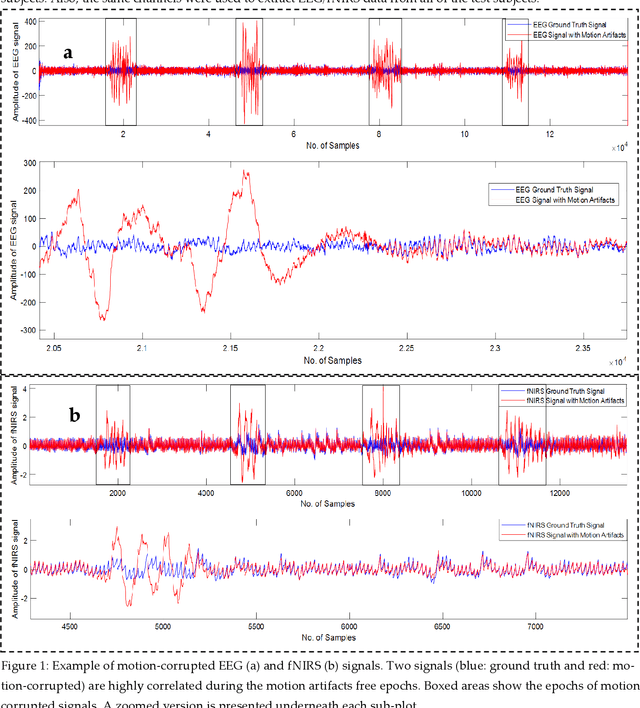
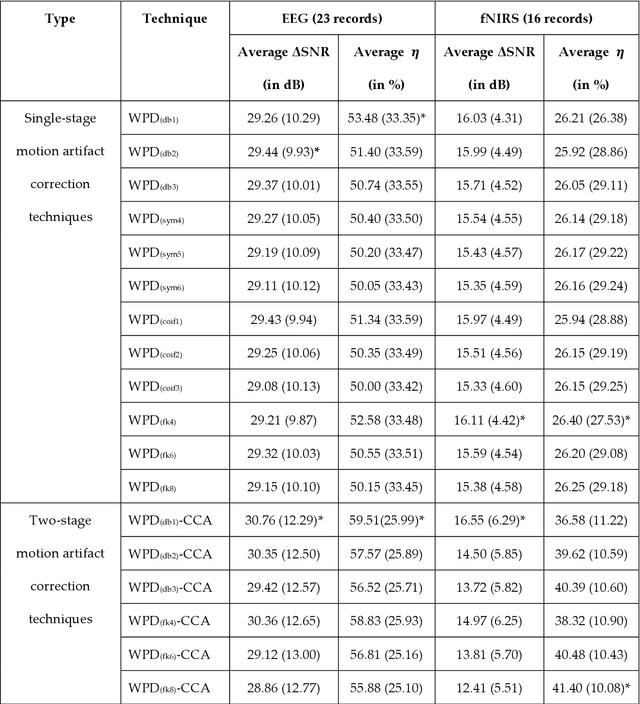
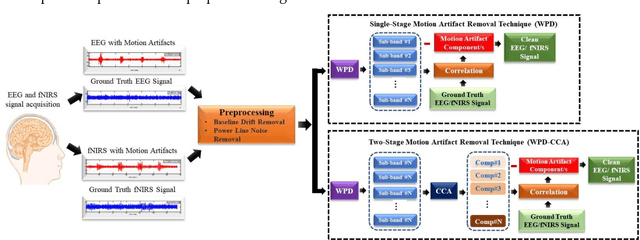
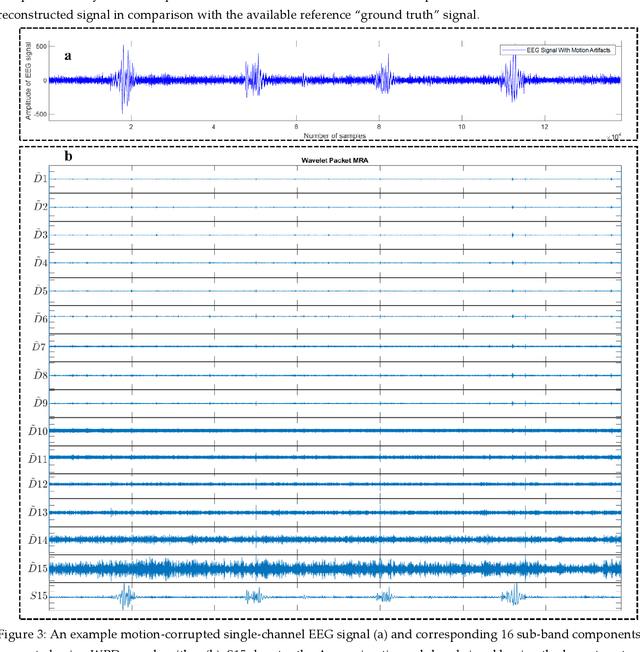
Abstract:The electroencephalogram (EEG) and functional near-infrared spectroscopy (fNIRS) signals, highly non-stationary in nature, greatly suffers from motion artifacts while recorded using wearable sensors. This paper proposes two robust methods: i) Wavelet packet decomposition (WPD), and ii) WPD in combination with canonical correlation analysis (WPD-CCA), for motion artifact correction from single-channel EEG and fNIRS signals. The efficacy of these proposed techniques is tested using a benchmark dataset and the performance of the proposed methods is measured using two well-established performance matrices: i) Difference in the signal to noise ratio ({\Delta}SNR) and ii) Percentage reduction in motion artifacts ({\eta}). The proposed WPD-based single-stage motion artifacts correction technique produces the highest average {\Delta}SNR (29.44 dB) when db2 wavelet packet is incorporated whereas the greatest average {\eta} (53.48%) is obtained using db1 wavelet packet for all the available 23 EEG recordings. Our proposed two-stage motion artifacts correction technique i.e. the WPD-CCA method utilizing db1 wavelet packet has shown the best denoising performance producing an average {\Delta}SNR and {\eta} values of 30.76 dB and 59.51%, respectively for all the EEG recordings. On the other hand, the two-stage motion artifacts removal technique i.e. WPD-CCA has produced the best average {\Delta}SNR (16.55 dB, utilizing db1 wavelet packet) and largest average {\eta} (41.40%, using fk8 wavelet packet). The highest average {\Delta}SNR and {\eta} using single-stage artifacts removal techniques (WPD) are found as 16.11 dB and 26.40%, respectively for all the fNIRS signals using fk4 wavelet packet. In both EEG and fNIRS modalities, the percentage reduction in motion artifacts increases by 11.28% and 56.82%, respectively when two-stage WPD-CCA techniques are employed.
Blind ECG Restoration by Operational Cycle-GANs
Jan 29, 2022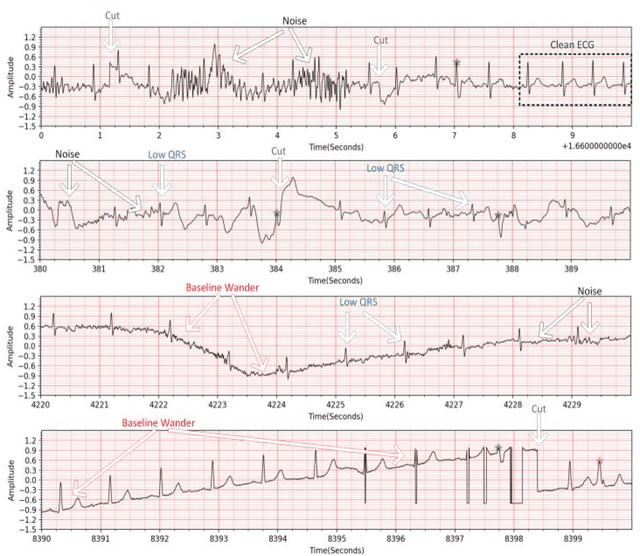
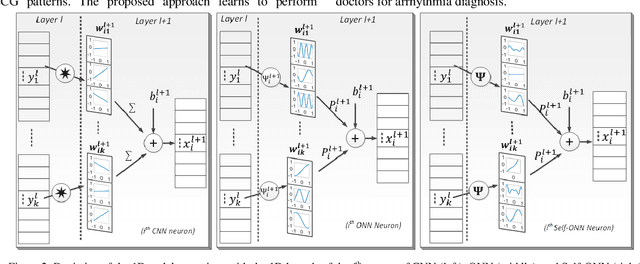
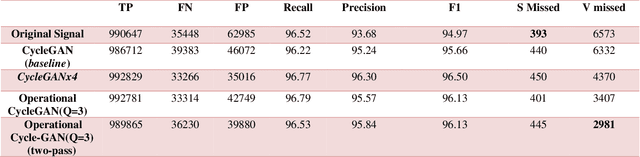
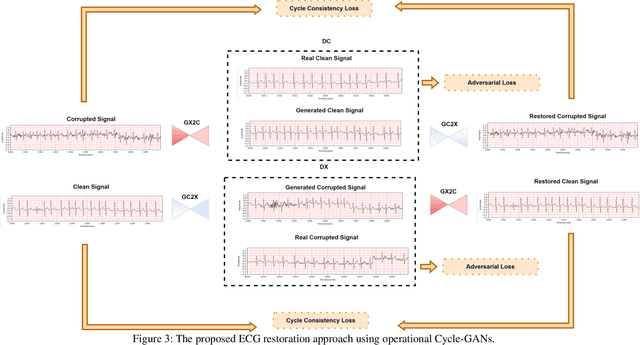
Abstract:Continuous long-term monitoring of electrocardiography (ECG) signals is crucial for the early detection of cardiac abnormalities such as arrhythmia. Non-clinical ECG recordings acquired by Holter and wearable ECG sensors often suffer from severe artifacts such as baseline wander, signal cuts, motion artifacts, variations on QRS amplitude, noise, and other interferences. Usually, a set of such artifacts occur on the same ECG signal with varying severity and duration, and this makes an accurate diagnosis by machines or medical doctors extremely difficult. Despite numerous studies that have attempted ECG denoising, they naturally fail to restore the actual ECG signal corrupted with such artifacts due to their simple and naive noise model. In this study, we propose a novel approach for blind ECG restoration using cycle-consistent generative adversarial networks (Cycle-GANs) where the quality of the signal can be improved to a clinical level ECG regardless of the type and severity of the artifacts corrupting the signal. To further boost the restoration performance, we propose 1D operational Cycle-GANs with the generative neuron model. The proposed approach has been evaluated extensively using one of the largest benchmark ECG datasets from the China Physiological Signal Challenge (CPSC-2020) with more than one million beats. Besides the quantitative and qualitative evaluations, a group of cardiologists performed medical evaluations to validate the quality and usability of the restored ECG, especially for an accurate arrhythmia diagnosis.
RamanNet: A generalized neural network architecture for Raman Spectrum Analysis
Jan 20, 2022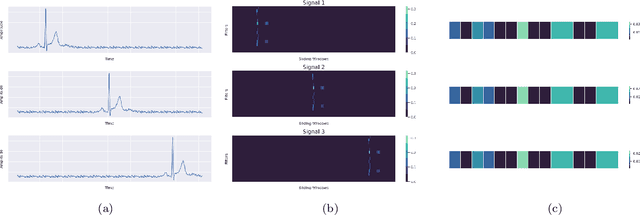
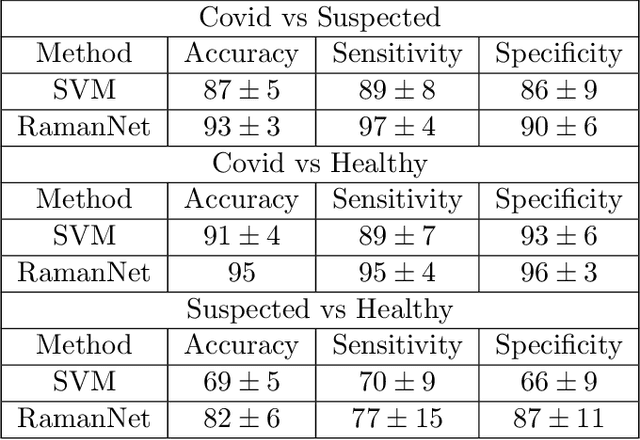
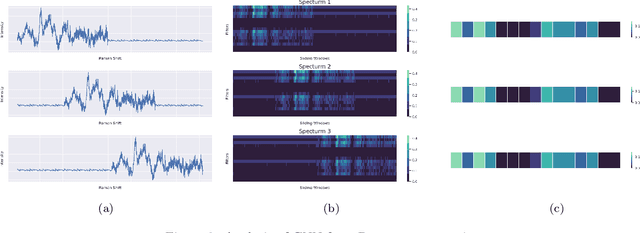
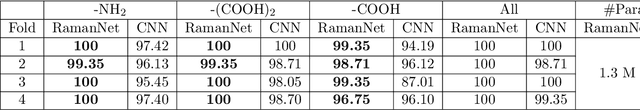
Abstract:Raman spectroscopy provides a vibrational profile of the molecules and thus can be used to uniquely identify different kind of materials. This sort of fingerprinting molecules has thus led to widespread application of Raman spectrum in various fields like medical dignostics, forensics, mineralogy, bacteriology and virology etc. Despite the recent rise in Raman spectra data volume, there has not been any significant effort in developing generalized machine learning methods for Raman spectra analysis. We examine, experiment and evaluate existing methods and conjecture that neither current sequential models nor traditional machine learning models are satisfactorily sufficient to analyze Raman spectra. Both has their perks and pitfalls, therefore we attempt to mix the best of both worlds and propose a novel network architecture RamanNet. RamanNet is immune to invariance property in CNN and at the same time better than traditional machine learning models for the inclusion of sparse connectivity. Our experiments on 4 public datasets demonstrate superior performance over the much complex state-of-the-art methods and thus RamanNet has the potential to become the defacto standard in Raman spectra data analysis
A Shallow U-Net Architecture for Reliably Predicting Blood Pressure from Photoplethysmogram and Electrocardiogram Signals
Nov 12, 2021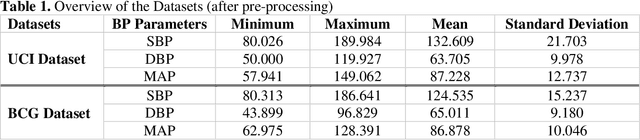
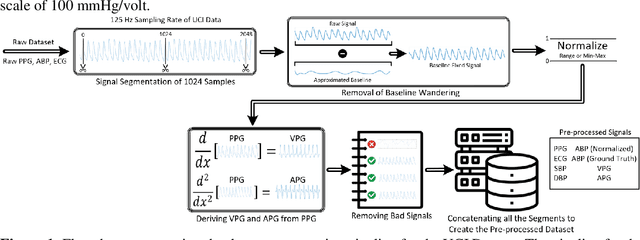
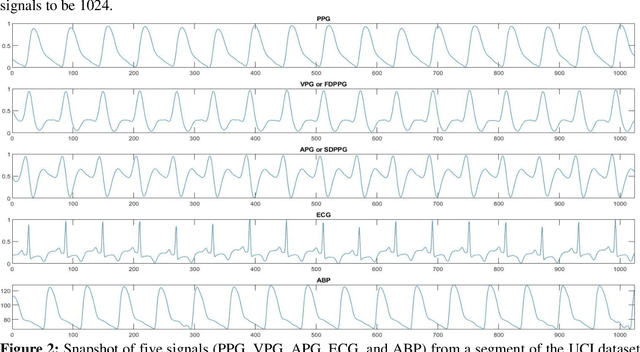
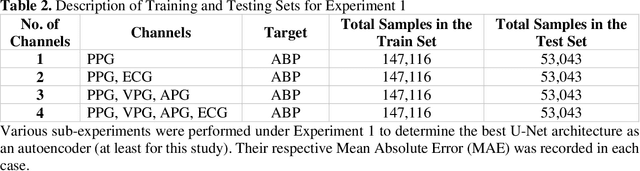
Abstract:Cardiovascular diseases are the most common causes of death around the world. To detect and treat heart-related diseases, continuous Blood Pressure (BP) monitoring along with many other parameters are required. Several invasive and non-invasive methods have been developed for this purpose. Most existing methods used in the hospitals for continuous monitoring of BP are invasive. On the contrary, cuff-based BP monitoring methods, which can predict Systolic Blood Pressure (SBP) and Diastolic Blood Pressure (DBP), cannot be used for continuous monitoring. Several studies attempted to predict BP from non-invasively collectible signals such as Photoplethysmogram (PPG) and Electrocardiogram (ECG), which can be used for continuous monitoring. In this study, we explored the applicability of autoencoders in predicting BP from PPG and ECG signals. The investigation was carried out on 12,000 instances of 942 patients of the MIMIC-II dataset and it was found that a very shallow, one-dimensional autoencoder can extract the relevant features to predict the SBP and DBP with the state-of-the-art performance on a very large dataset. Independent test set from a portion of the MIMIC-II dataset provides an MAE of 2.333 and 0.713 for SBP and DBP, respectively. On an external dataset of forty subjects, the model trained on the MIMIC-II dataset, provides an MAE of 2.728 and 1.166 for SBP and DBP, respectively. For both the cases, the results met British Hypertension Society (BHS) Grade A and surpassed the studies from the current literature.
Robust Peak Detection for Holter ECGs by Self-Organized Operational Neural Networks
Sep 30, 2021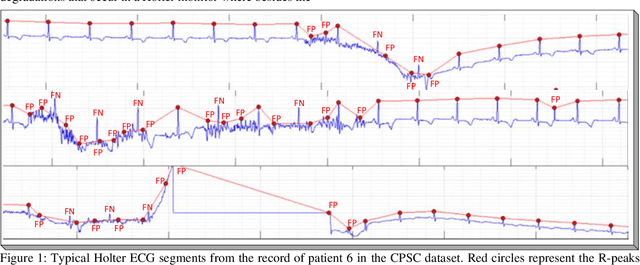
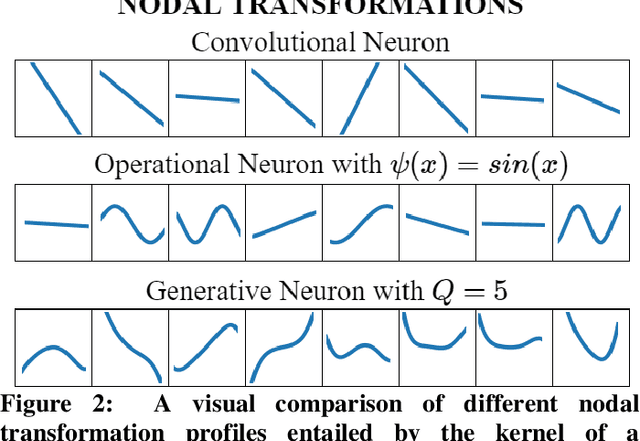
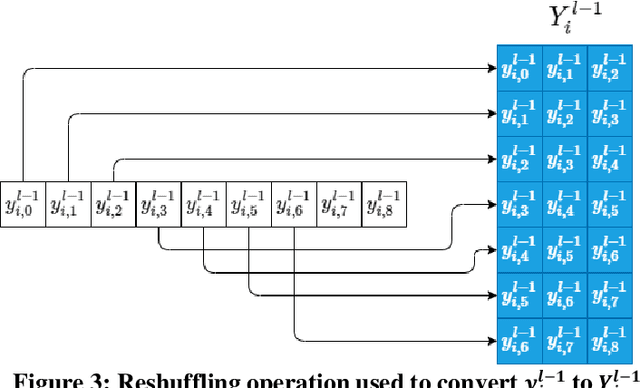
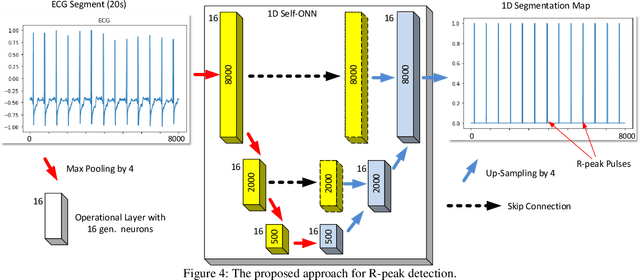
Abstract:Although numerous R-peak detectors have been proposed in the literature, their robustness and performance levels may significantly deteriorate in low quality and noisy signals acquired from mobile ECG sensors such as Holter monitors. Recently, this issue has been addressed by deep 1D Convolutional Neural Networks (CNNs) that have achieved state-of-the-art performance levels in Holter monitors; however, they pose a high complexity level that requires special parallelized hardware setup for real-time processing. On the other hand, their performance deteriorates when a compact network configuration is used instead. This is an expected outcome as recent studies have demonstrated that the learning performance of CNNs is limited due to their strictly homogenous configuration with the sole linear neuron model. This has been addressed by Operational Neural Networks (ONNs) with their heterogenous network configuration encapsulating neurons with various non-linear operators. In this study, to further boost the peak detection performance along with an elegant computational efficiency, we propose 1D Self-Organized Operational Neural Networks (Self-ONNs) with generative neurons. The most crucial advantage of 1D Self-ONNs over the ONNs is their self-organization capability that voids the need to search for the best operator set per neuron since each generative neuron has the ability to create the optimal operator during training. The experimental results over the China Physiological Signal Challenge-2020 (CPSC) dataset with more than one million ECG beats show that the proposed 1D Self-ONNs can significantly surpass the state-of-the-art deep CNN with less computational complexity. Results demonstrate that the proposed solution achieves 99.10% F1-score, 99.79% sensitivity, and 98.42% positive predictivity in the CPSC dataset which is the best R-peak detection performance ever achieved.
A Machine Learning Model for Early Detection of Diabetic Foot using Thermogram Images
Jun 27, 2021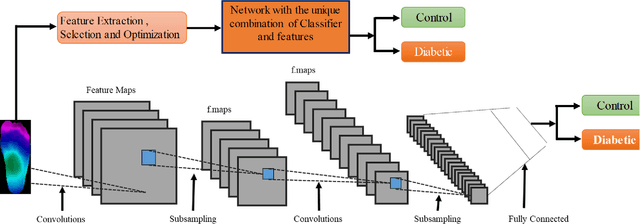
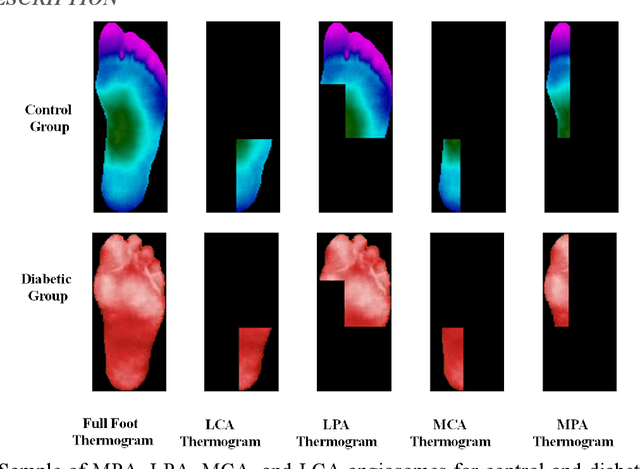
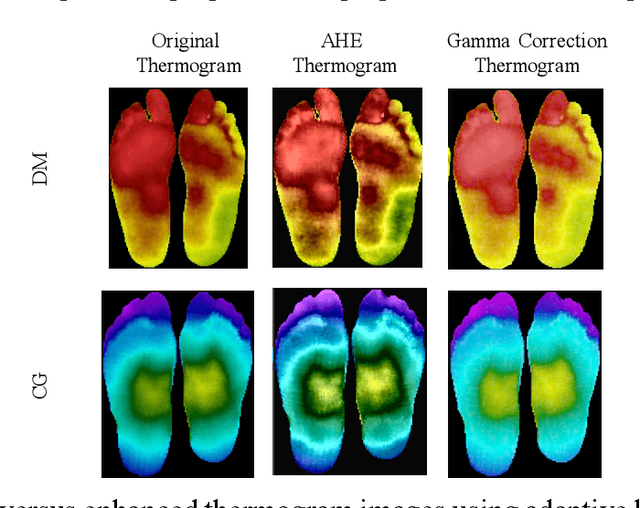
Abstract:Diabetes foot ulceration (DFU) and amputation are a cause of significant morbidity. The prevention of DFU may be achieved by the identification of patients at risk of DFU and the institution of preventative measures through education and offloading. Several studies have reported that thermogram images may help to detect an increase in plantar temperature prior to DFU. However, the distribution of plantar temperature may be heterogeneous, making it difficult to quantify and utilize to predict outcomes. We have compared a machine learning-based scoring technique with feature selection and optimization techniques and learning classifiers to several state-of-the-art Convolutional Neural Networks (CNNs) on foot thermogram images and propose a robust solution to identify the diabetic foot. A comparatively shallow CNN model, MobilenetV2 achieved an F1 score of ~95% for a two-feet thermogram image-based classification and the AdaBoost Classifier used 10 features and achieved an F1 score of 97 %. A comparison of the inference time for the best-performing networks confirmed that the proposed algorithm can be deployed as a smartphone application to allow the user to monitor the progression of the DFU in a home setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge