Alexander Geiger
On the notion of missingness for path attribution explainability methods in medical settings: Guiding the selection of medically meaningful baselines
Aug 20, 2025Abstract:The explainability of deep learning models remains a significant challenge, particularly in the medical domain where interpretable outputs are critical for clinical trust and transparency. Path attribution methods such as Integrated Gradients rely on a baseline input representing the absence of relevant features ("missingness"). Commonly used baselines, such as all-zero inputs, are often semantically meaningless, especially in medical contexts where missingness can itself be informative. While alternative baseline choices have been explored, existing methods lack a principled approach to dynamically select baselines tailored to each input. In this work, we examine the notion of missingness in the medical setting, analyze its implications for baseline selection, and introduce a counterfactual-guided approach to address the limitations of conventional baselines. We argue that a clinically normal but input-close counterfactual represents a more accurate representation of a meaningful absence of features in medical data. To implement this, we use a Variational Autoencoder to generate counterfactual baselines, though our concept is generative-model-agnostic and can be applied with any suitable counterfactual method. We evaluate the approach on three distinct medical data sets and empirically demonstrate that counterfactual baselines yield more faithful and medically relevant attributions compared to standard baseline choices.
Detecting and clustering swallow events in esophageal long-term high-resolution manometry
May 02, 2024



Abstract:High-resolution manometry (HRM) is the gold standard in diagnosing esophageal motility disorders. As HRM is typically conducted under short-term laboratory settings, intermittently occurring disorders are likely to be missed. Therefore, long-term (up to 24h) HRM (LTHRM) is used to gain detailed insights into the swallowing behavior. However, analyzing the extensive data from LTHRM is challenging and time consuming as medical experts have to analyze the data manually, which is slow and prone to errors. To address this challenge, we propose a Deep Learning based swallowing detection method to accurately identify swallowing events and secondary non-deglutitive-induced esophageal motility disorders in LTHRM data. We then proceed with clustering the identified swallows into distinct classes, which are analyzed by highly experienced clinicians to validate the different swallowing patterns. We evaluate our computational pipeline on a total of 25 LTHRMs, which were meticulously annotated by medical experts. By detecting more than 94% of all relevant swallow events and providing all relevant clusters for a more reliable diagnostic process among experienced clinicians, we are able to demonstrate the effectiveness as well as positive clinical impact of our approach to make LTHRM feasible in clinical care.
TadGAN: Time Series Anomaly Detection Using Generative Adversarial Networks
Sep 19, 2020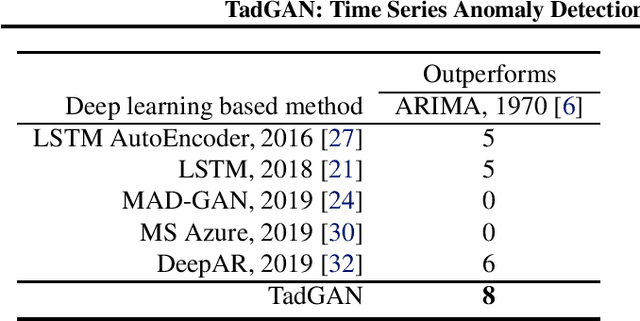
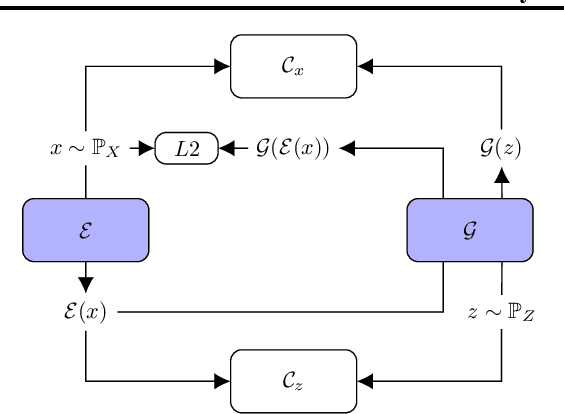
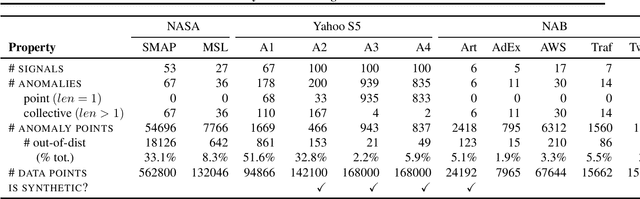
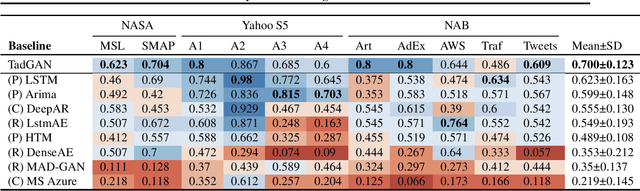
Abstract:Time series anomalies can offer information relevant to critical situations facing various fields, from finance and aerospace to the IT, security, and medical domains. However, detecting anomalies in time series data is particularly challenging due to the vague definition of anomalies and said data's frequent lack of labels and highly complex temporal correlations. Current state-of-the-art unsupervised machine learning methods for anomaly detection suffer from scalability and portability issues, and may have high false positive rates. In this paper, we propose TadGAN, an unsupervised anomaly detection approach built on Generative Adversarial Networks (GANs). To capture the temporal correlations of time series distributions, we use LSTM Recurrent Neural Networks as base models for Generators and Critics. TadGAN is trained with cycle consistency loss to allow for effective time-series data reconstruction. We further propose several novel methods to compute reconstruction errors, as well as different approaches to combine reconstruction errors and Critic outputs to compute anomaly scores. To demonstrate the performance and generalizability of our approach, we test several anomaly scoring techniques and report the best-suited one. We compare our approach to 8 baseline anomaly detection methods on 11 datasets from multiple reputable sources such as NASA, Yahoo, Numenta, Amazon, and Twitter. The results show that our approach can effectively detect anomalies and outperform baseline methods in most cases (6 out of 11). Notably, our method has the highest averaged F1 score across all the datasets. Our code is open source and is available as a benchmarking tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge