Alan Evans
EEG Foundation Challenge: From Cross-Task to Cross-Subject EEG Decoding
Jun 23, 2025Abstract:Current electroencephalogram (EEG) decoding models are typically trained on small numbers of subjects performing a single task. Here, we introduce a large-scale, code-submission-based competition comprising two challenges. First, the Transfer Challenge asks participants to build and test a model that can zero-shot decode new tasks and new subjects from their EEG data. Second, the Psychopathology factor prediction Challenge asks participants to infer subject measures of mental health from EEG data. For this, we use an unprecedented, multi-terabyte dataset of high-density EEG signals (128 channels) recorded from over 3,000 child to young adult subjects engaged in multiple active and passive tasks. We provide several tunable neural network baselines for each of these two challenges, including a simple network and demographic-based regression models. Developing models that generalise across tasks and individuals will pave the way for ML network architectures capable of adapting to EEG data collected from diverse tasks and individuals. Similarly, predicting mental health-relevant personality trait values from EEG might identify objective biomarkers useful for clinical diagnosis and design of personalised treatment for psychological conditions. Ultimately, the advances spurred by this challenge could contribute to the development of computational psychiatry and useful neurotechnology, and contribute to breakthroughs in both fundamental neuroscience and applied clinical research.
* Approved at Neurips Competition track. webpage: https://eeg2025.github.io/
Confidence-Guided Unsupervised Domain Adaptation for Cerebellum Segmentation
Jun 14, 2022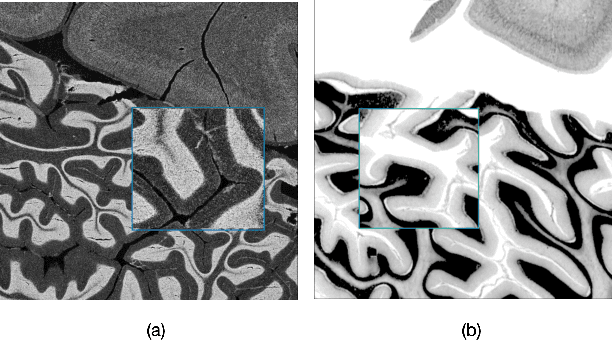
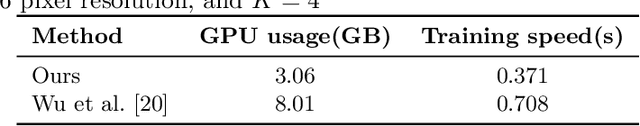
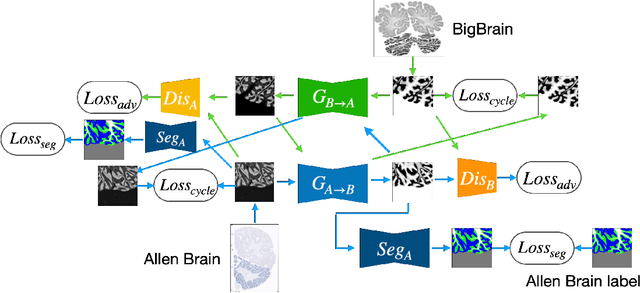
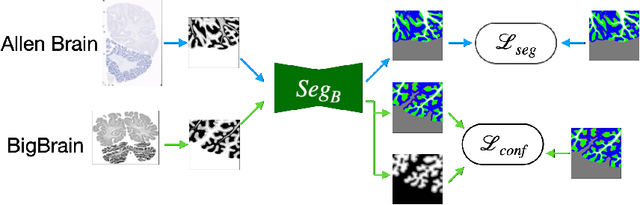
Abstract:The lack of a comprehensive high-resolution atlas of the cerebellum has hampered studies of cerebellar involvement in normal brain function and disease. A good representation of the tightly foliated aspect of the cerebellar cortex is difficult to achieve because of the highly convoluted surface and the time it would take for manual delineation. The quality of manual segmentation is influenced by human expert judgment, and automatic labelling is constrained by the limited robustness of existing segmentation algorithms. The 20umisotropic BigBrain dataset provides an unprecedented high resolution framework for semantic segmentation compared to the 1000um(1mm) resolution afforded by magnetic resonance imaging. To dispense with the manual annotation requirement, we propose to train a model to adaptively transfer the annotation from the cerebellum on the Allen Brain Human Brain Atlas to the BigBrain in an unsupervised manner, taking into account the different staining and spacing between sections. The distinct visual discrepancy between the Allen Brain and BigBrain prevents existing approaches to provide meaningful segmentation masks, and artifacts caused by sectioning and histological slice preparation in the BigBrain data pose an extra challenge. To address these problems, we propose a two-stage framework where we first transfer the Allen Brain cerebellum to a space sharing visual similarity with the BigBrain. We then introduce a self-training strategy with a confidence map to guide the model learning from the noisy pseudo labels iteratively. Qualitative results validate the effectiveness of our approach, and quantitative experiments reveal that our method can achieve over 2.6% loss reduction compared with other approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge