Deep Learning Surrogate for Fast CIR Prediction in Reactive Molecular Diffusion Advection Channels
Paper and Code
Dec 19, 2025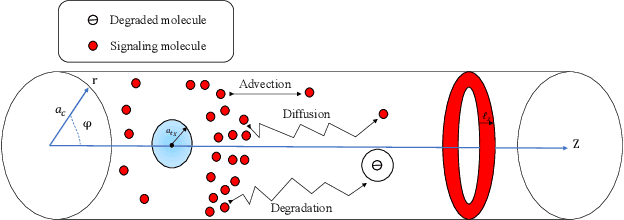
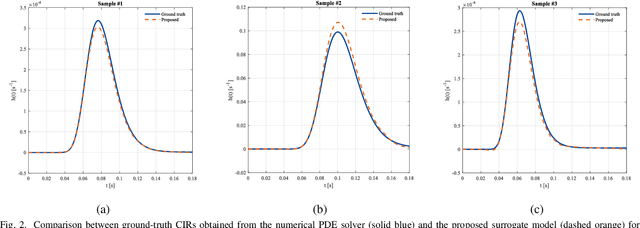
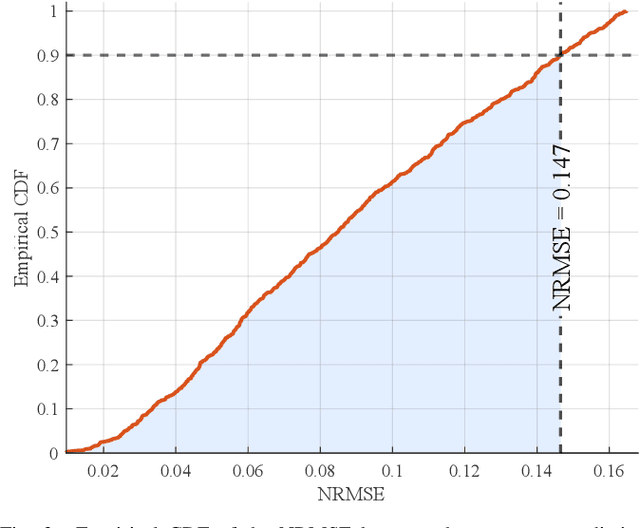
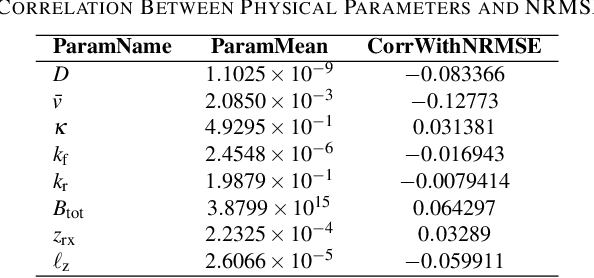
Accurate channel impulse response (CIR) modeling in molecular communication (MC) often requires solving coupled reactive diffusion-advection equations, which is computationally expensive for large parameter sweeps or design loops. We develop a deep-learning surrogate for a three-dimensional duct MC channel with reactive diffusion-advection transport and reversible ligand-receptor binding on a finite ring receiver. Using a physics-based partial differential equation (PDE)-ordinary differential equation (ODE) model, we generate a large CIR dataset across broad transport, reaction, and geometric ranges and train a neural network that maps these parameters directly to the CIR. On an independent test set, the surrogate closely matches reference CIRs both qualitatively and quantitatively: the empirical cumulative distribution function (CDF) of the normalized root mean square error (NRMSE) shows that 90% of test channels are predicted with error below 0.15, with only weak dependence on individual parameters. The surrogate therefore offers an accurate and computationally efficient replacement for repeated PDE-based CIR evaluations in MC system analysis and design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge