Deep learning-derived arterial input function
Paper and Code
May 30, 2025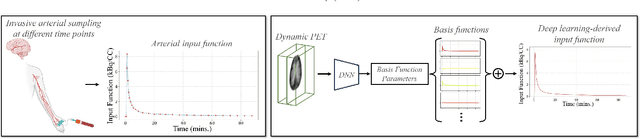
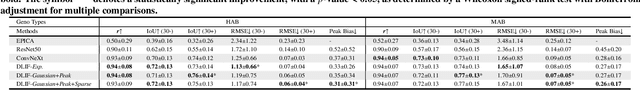
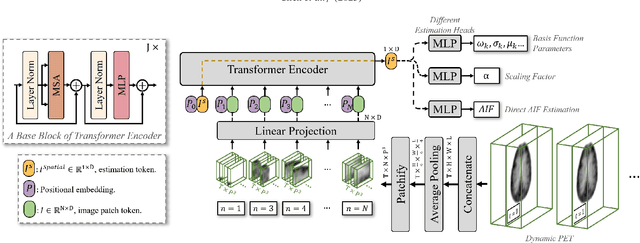
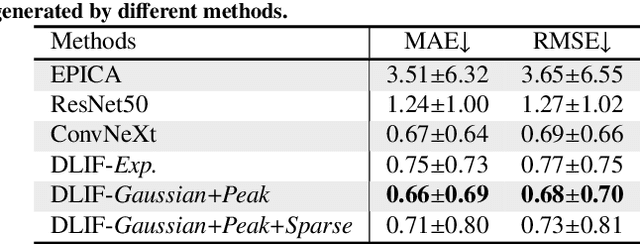
Dynamic positron emission tomography (PET) imaging combined with radiotracer kinetic modeling is a powerful technique for visualizing biological processes in the brain, offering valuable insights into brain functions and neurological disorders such as Alzheimer's and Parkinson's diseases. Accurate kinetic modeling relies heavily on the use of a metabolite-corrected arterial input function (AIF), which typically requires invasive and labor-intensive arterial blood sampling. While alternative non-invasive approaches have been proposed, they often compromise accuracy or still necessitate at least one invasive blood sampling. In this study, we present the deep learning-derived arterial input function (DLIF), a deep learning framework capable of estimating a metabolite-corrected AIF directly from dynamic PET image sequences without any blood sampling. We validated DLIF using existing dynamic PET patient data. We compared DLIF and resulting parametric maps against ground truth measurements. Our evaluation shows that DLIF achieves accurate and robust AIF estimation. By leveraging deep learning's ability to capture complex temporal dynamics and incorporating prior knowledge of typical AIF shapes through basis functions, DLIF provides a rapid, accurate, and entirely non-invasive alternative to traditional AIF measurement methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge