Zhiyu Wan
A DICOM Image De-identification Algorithm in the MIDI-B Challenge
Aug 11, 2025Abstract:Image de-identification is essential for the public sharing of medical images, particularly in the widely used Digital Imaging and Communications in Medicine (DICOM) format as required by various regulations and standards, including Health Insurance Portability and Accountability Act (HIPAA) privacy rules, the DICOM PS3.15 standard, and best practices recommended by the Cancer Imaging Archive (TCIA). The Medical Image De-Identification Benchmark (MIDI-B) Challenge at the 27th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2024) was organized to evaluate rule-based DICOM image de-identification algorithms with a large dataset of clinical DICOM images. In this report, we explore the critical challenges of de-identifying DICOM images, emphasize the importance of removing personally identifiable information (PII) to protect patient privacy while ensuring the continued utility of medical data for research, diagnostics, and treatment, and provide a comprehensive overview of the standards and regulations that govern this process. Additionally, we detail the de-identification methods we applied - such as pixel masking, date shifting, date hashing, text recognition, text replacement, and text removal - to process datasets during the test phase in strict compliance with these standards. According to the final leaderboard of the MIDI-B challenge, the latest version of our solution algorithm correctly executed 99.92% of the required actions and ranked 2nd out of 10 teams that completed the challenge (from a total of 22 registered teams). Finally, we conducted a thorough analysis of the resulting statistics and discussed the limitations of current approaches and potential avenues for future improvement.
Medical Image De-Identification Benchmark Challenge
Jul 31, 2025



Abstract:The de-identification (deID) of protected health information (PHI) and personally identifiable information (PII) is a fundamental requirement for sharing medical images, particularly through public repositories, to ensure compliance with patient privacy laws. In addition, preservation of non-PHI metadata to inform and enable downstream development of imaging artificial intelligence (AI) is an important consideration in biomedical research. The goal of MIDI-B was to provide a standardized platform for benchmarking of DICOM image deID tools based on a set of rules conformant to the HIPAA Safe Harbor regulation, the DICOM Attribute Confidentiality Profiles, and best practices in preservation of research-critical metadata, as defined by The Cancer Imaging Archive (TCIA). The challenge employed a large, diverse, multi-center, and multi-modality set of real de-identified radiology images with synthetic PHI/PII inserted. The MIDI-B Challenge consisted of three phases: training, validation, and test. Eighty individuals registered for the challenge. In the training phase, we encouraged participants to tune their algorithms using their in-house or public data. The validation and test phases utilized the DICOM images containing synthetic identifiers (of 216 and 322 subjects, respectively). Ten teams successfully completed the test phase of the challenge. To measure success of a rule-based approach to image deID, scores were computed as the percentage of correct actions from the total number of required actions. The scores ranged from 97.91% to 99.93%. Participants employed a variety of open-source and proprietary tools with customized configurations, large language models, and optical character recognition (OCR). In this paper we provide a comprehensive report on the MIDI-B Challenge's design, implementation, results, and lessons learned.
A Multifaceted Benchmarking of Synthetic Electronic Health Record Generation Models
Aug 02, 2022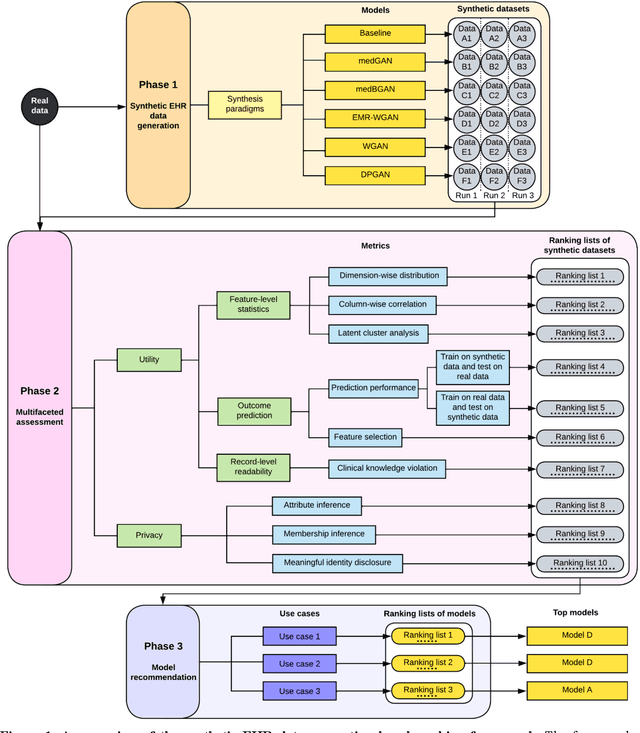
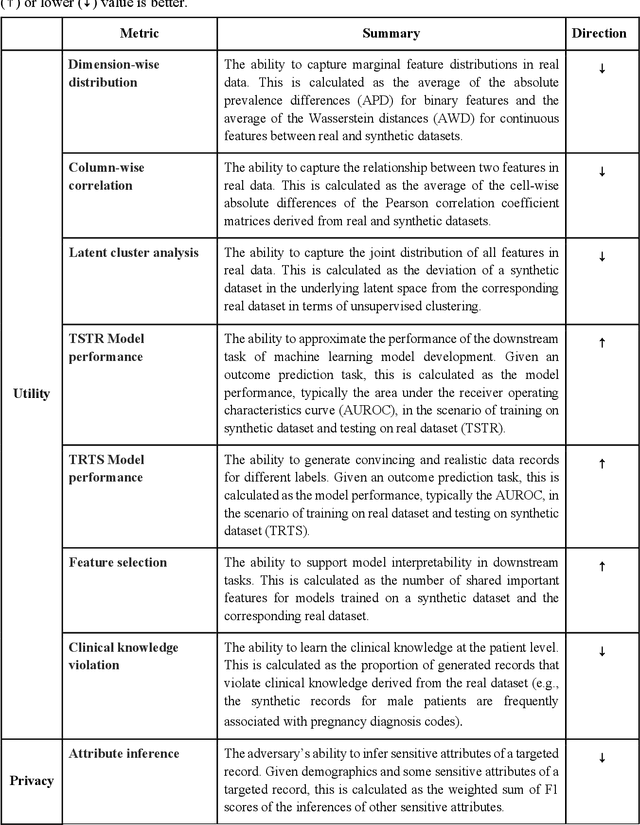
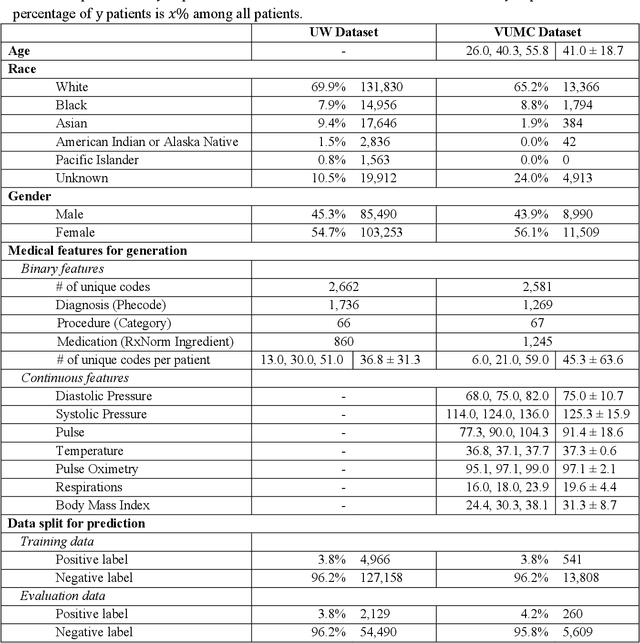
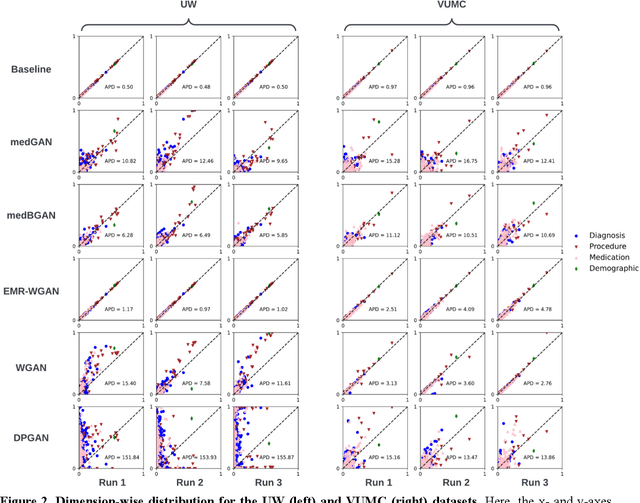
Abstract:Synthetic health data have the potential to mitigate privacy concerns when sharing data to support biomedical research and the development of innovative healthcare applications. Modern approaches for data generation based on machine learning, generative adversarial networks (GAN) methods in particular, continue to evolve and demonstrate remarkable potential. Yet there is a lack of a systematic assessment framework to benchmark methods as they emerge and determine which methods are most appropriate for which use cases. In this work, we introduce a generalizable benchmarking framework to appraise key characteristics of synthetic health data with respect to utility and privacy metrics. We apply the framework to evaluate synthetic data generation methods for electronic health records (EHRs) data from two large academic medical centers with respect to several use cases. The results illustrate that there is a utility-privacy tradeoff for sharing synthetic EHR data. The results further indicate that no method is unequivocally the best on all criteria in each use case, which makes it evident why synthetic data generation methods need to be assessed in context.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge