Ying Xia
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025



Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Automated volumetric and statistical shape assessment of cam-type morphology of the femoral head-neck region from 3D magnetic resonance images
Dec 06, 2021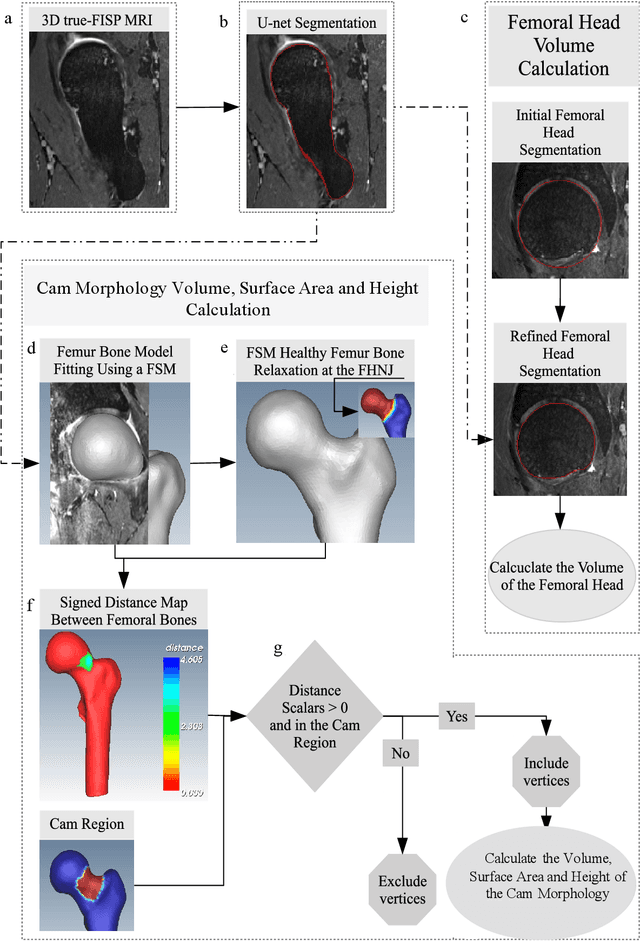
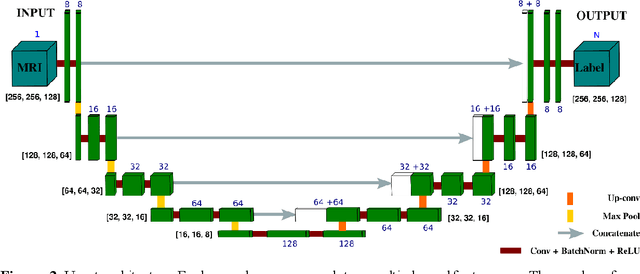

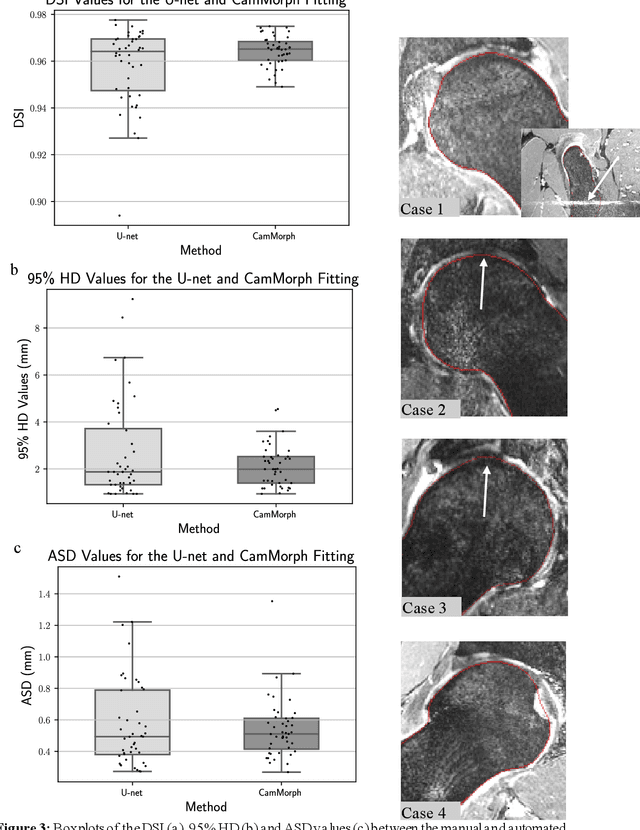
Abstract:Femoroacetabular impingement (FAI) cam morphology is routinely assessed using two-dimensional alpha angles which do not provide specific data on cam size characteristics. The purpose of this study is to implement a novel, automated three-dimensional (3D) pipeline, CamMorph, for segmentation and measurement of cam volume, surface area and height from magnetic resonance (MR) images in patients with FAI. The CamMorph pipeline involves two processes: i) proximal femur segmentation using an approach integrating 3D U-net with focused shape modelling (FSM); ii) use of patient-specific anatomical information from 3D FSM to simulate healthy femoral bone models and pathological region constraints to identify cam bone mass. Agreement between manual and automated segmentation of the proximal femur was evaluated with the Dice similarity index (DSI) and surface distance measures. Independent t-tests or Mann-Whitney U rank tests were used to compare the femoral head volume, cam volume, surface area and height data between female and male patients with FAI. There was a mean DSI value of 0.964 between manual and automated segmentation of proximal femur volume. Compared to female FAI patients, male patients had a significantly larger mean femoral head volume (66.12cm3 v 46.02cm3, p<0.001). Compared to female FAI patients, male patients had a significantly larger mean cam volume (1136.87mm3 v 337.86mm3, p<0.001), surface area (657.36mm2 v 306.93mm2 , p<0.001), maximum-height (3.89mm v 2.23mm, p<0.001) and average-height (1.94mm v 1.00mm, p<0.001). Automated analyses of 3D MR images from patients with FAI using the CamMorph pipeline showed that, in comparison with female patients, male patients had significantly greater cam volume, surface area and height.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge