Xiaolin Cheng
GeoPep: A geometry-aware masked language model for protein-peptide binding site prediction
Oct 30, 2025Abstract:Multimodal approaches that integrate protein structure and sequence have achieved remarkable success in protein-protein interface prediction. However, extending these methods to protein-peptide interactions remains challenging due to the inherent conformational flexibility of peptides and the limited availability of structural data that hinder direct training of structure-aware models. To address these limitations, we introduce GeoPep, a novel framework for peptide binding site prediction that leverages transfer learning from ESM3, a multimodal protein foundation model. GeoPep fine-tunes ESM3's rich pre-learned representations from protein-protein binding to address the limited availability of protein-peptide binding data. The fine-tuned model is further integrated with a parameter-efficient neural network architecture capable of learning complex patterns from sparse data. Furthermore, the model is trained using distance-based loss functions that exploit 3D structural information to enhance binding site prediction. Comprehensive evaluations demonstrate that GeoPep significantly outperforms existing methods in protein-peptide binding site prediction by effectively capturing sparse and heterogeneous binding patterns.
Target specific peptide design using latent space approximate trajectory collector
Feb 02, 2023



Abstract:Despite the prevalence and many successes of deep learning applications in de novo molecular design, the problem of peptide generation targeting specific proteins remains unsolved. A main barrier for this is the scarcity of the high-quality training data. To tackle the issue, we propose a novel machine learning based peptide design architecture, called Latent Space Approximate Trajectory Collector (LSATC). It consists of a series of samplers on an optimization trajectory on a highly non-convex energy landscape that approximates the distributions of peptides with desired properties in a latent space. The process involves little human intervention and can be implemented in an end-to-end manner. We demonstrate the model by the design of peptide extensions targeting Beta-catenin, a key nuclear effector protein involved in canonical Wnt signalling. When compared with a random sampler, LSATC can sample peptides with $36\%$ lower binding scores in a $16$ times smaller interquartile range (IQR) and $284\%$ less hydrophobicity with a $1.4$ times smaller IQR. LSATC also largely outperforms other common generative models. Finally, we utilized a clustering algorithm to select 4 peptides from the 100 LSATC designed peptides for experimental validation. The result confirms that all the four peptides extended by LSATC show improved Beta-catenin binding by at least $20.0\%$, and two of the peptides show a $3$ fold increase in binding affinity as compared to the base peptide.
Compiler-Level Matrix Multiplication Optimization for Deep Learning
Sep 23, 2019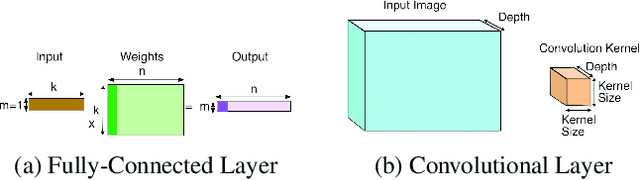
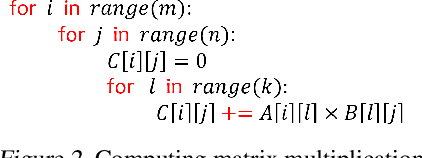
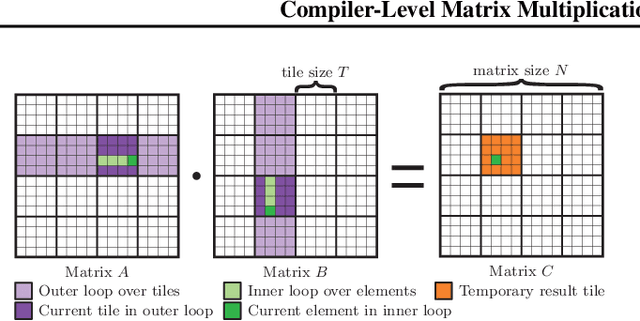

Abstract:An important linear algebra routine, GEneral Matrix Multiplication (GEMM), is a fundamental operator in deep learning. Compilers need to translate these routines into low-level code optimized for specific hardware. Compiler-level optimization of GEMM has significant performance impact on training and executing deep learning models. However, most deep learning frameworks rely on hardware-specific operator libraries in which GEMM optimization has been mostly achieved by manual tuning, which restricts the performance on different target hardware. In this paper, we propose two novel algorithms for GEMM optimization based on the TVM framework, a lightweight Greedy Best First Search (G-BFS) method based on heuristic search, and a Neighborhood Actor Advantage Critic (N-A2C) method based on reinforcement learning. Experimental results show significant performance improvement of the proposed methods, in both the optimality of the solution and the cost of search in terms of time and fraction of the search space explored. Specifically, the proposed methods achieve 24% and 40% savings in GEMM computation time over state-of-the-art XGBoost and RNN methods, respectively, while exploring only 0.1% of the search space. The proposed approaches have potential to be applied to other operator-level optimizations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge