Vivek Chawla
DIVIDE: A Framework for Learning from Independent Multi-Mechanism Data Using Deep Encoders and Gaussian Processes
Nov 16, 2025Abstract:Scientific datasets often arise from multiple independent mechanisms such as spatial, categorical or structural effects, whose combined influence obscures their individual contributions. We introduce DIVIDE, a framework that disentangles these influences by integrating mechanism-specific deep encoders with a structured Gaussian Process in a joint latent space. Disentanglement here refers to separating independently acting generative factors. The encoders isolate distinct mechanisms while the Gaussian Process captures their combined effect with calibrated uncertainty. The architecture supports structured priors, enabling interpretable and mechanism-aware prediction as well as efficient active learning. DIVIDE is demonstrated on synthetic datasets combining categorical image patches with nonlinear spatial fields, on FerroSIM spin lattice simulations of ferroelectric patterns, and on experimental PFM hysteresis loops from PbTiO3 films. Across benchmarks, DIVIDE separates mechanisms, reproduces additive and scaled interactions, and remains robust under noise. The framework extends naturally to multifunctional datasets where mechanical, electromagnetic or optical responses coexist.
Mic-hackathon 2024: Hackathon on Machine Learning for Electron and Scanning Probe Microscopy
Jun 10, 2025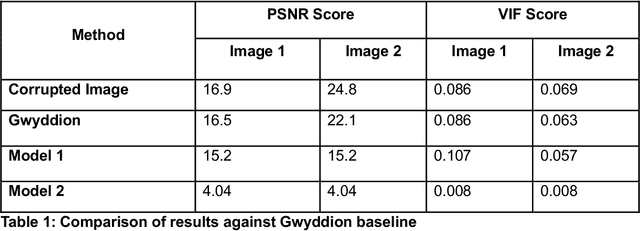
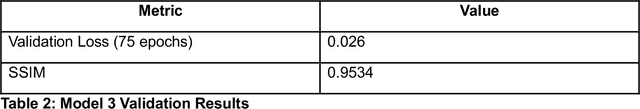
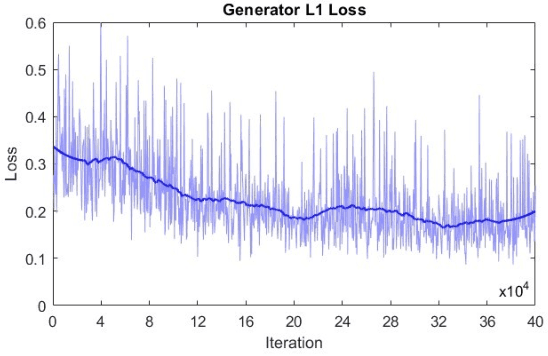
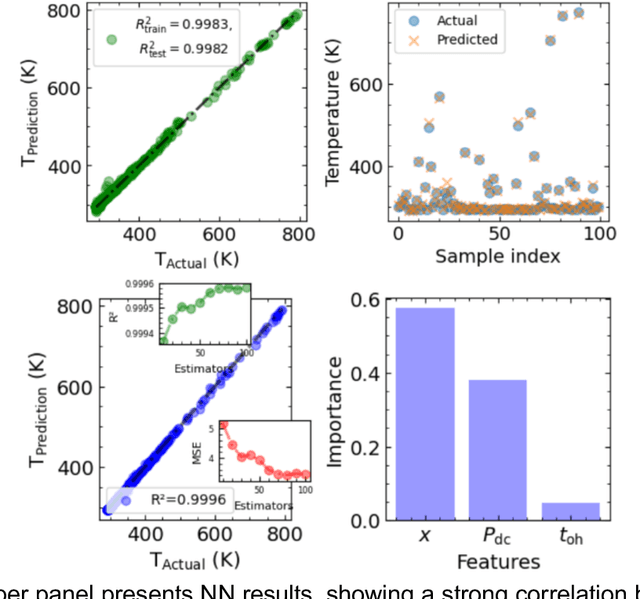
Abstract:Microscopy is a primary source of information on materials structure and functionality at nanometer and atomic scales. The data generated is often well-structured, enriched with metadata and sample histories, though not always consistent in detail or format. The adoption of Data Management Plans (DMPs) by major funding agencies promotes preservation and access. However, deriving insights remains difficult due to the lack of standardized code ecosystems, benchmarks, and integration strategies. As a result, data usage is inefficient and analysis time is extensive. In addition to post-acquisition analysis, new APIs from major microscope manufacturers enable real-time, ML-based analytics for automated decision-making and ML-agent-controlled microscope operation. Yet, a gap remains between the ML and microscopy communities, limiting the impact of these methods on physics, materials discovery, and optimization. Hackathons help bridge this divide by fostering collaboration between ML researchers and microscopy experts. They encourage the development of novel solutions that apply ML to microscopy, while preparing a future workforce for instrumentation, materials science, and applied ML. This hackathon produced benchmark datasets and digital twins of microscopes to support community growth and standardized workflows. All related code is available at GitHub: https://github.com/KalininGroup/Mic-hackathon-2024-codes-publication/tree/1.0.0.1
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge