Zhaslan Baraissov
Mic-hackathon 2024: Hackathon on Machine Learning for Electron and Scanning Probe Microscopy
Jun 10, 2025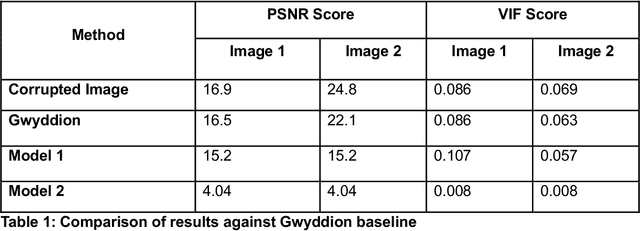
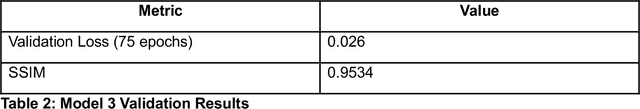
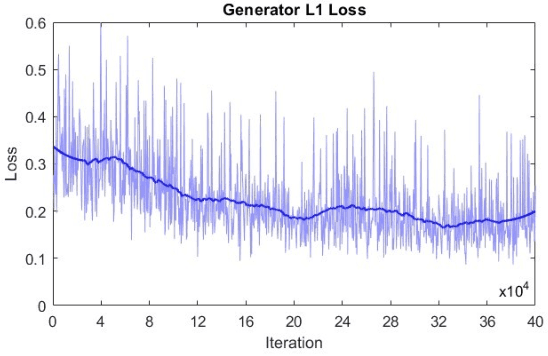
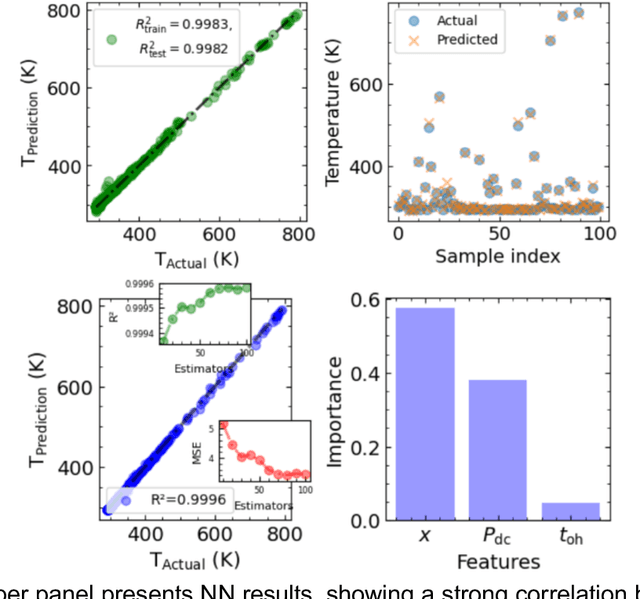
Abstract:Microscopy is a primary source of information on materials structure and functionality at nanometer and atomic scales. The data generated is often well-structured, enriched with metadata and sample histories, though not always consistent in detail or format. The adoption of Data Management Plans (DMPs) by major funding agencies promotes preservation and access. However, deriving insights remains difficult due to the lack of standardized code ecosystems, benchmarks, and integration strategies. As a result, data usage is inefficient and analysis time is extensive. In addition to post-acquisition analysis, new APIs from major microscope manufacturers enable real-time, ML-based analytics for automated decision-making and ML-agent-controlled microscope operation. Yet, a gap remains between the ML and microscopy communities, limiting the impact of these methods on physics, materials discovery, and optimization. Hackathons help bridge this divide by fostering collaboration between ML researchers and microscopy experts. They encourage the development of novel solutions that apply ML to microscopy, while preparing a future workforce for instrumentation, materials science, and applied ML. This hackathon produced benchmark datasets and digital twins of microscopes to support community growth and standardized workflows. All related code is available at GitHub: https://github.com/KalininGroup/Mic-hackathon-2024-codes-publication/tree/1.0.0.1
Single-shot pop-out 3D metrology of thin specimens with TEM
Sep 07, 2022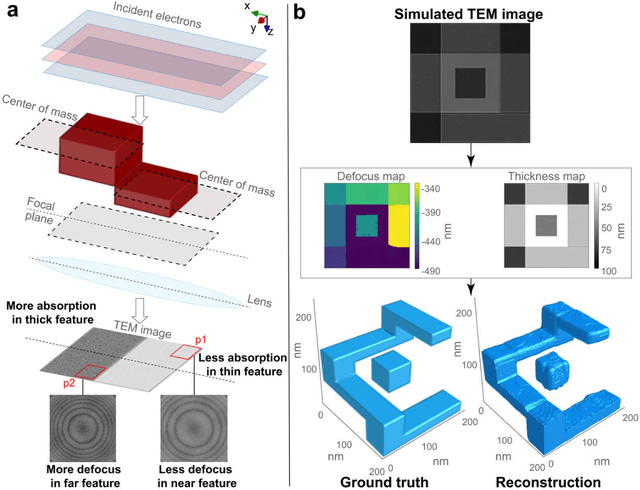
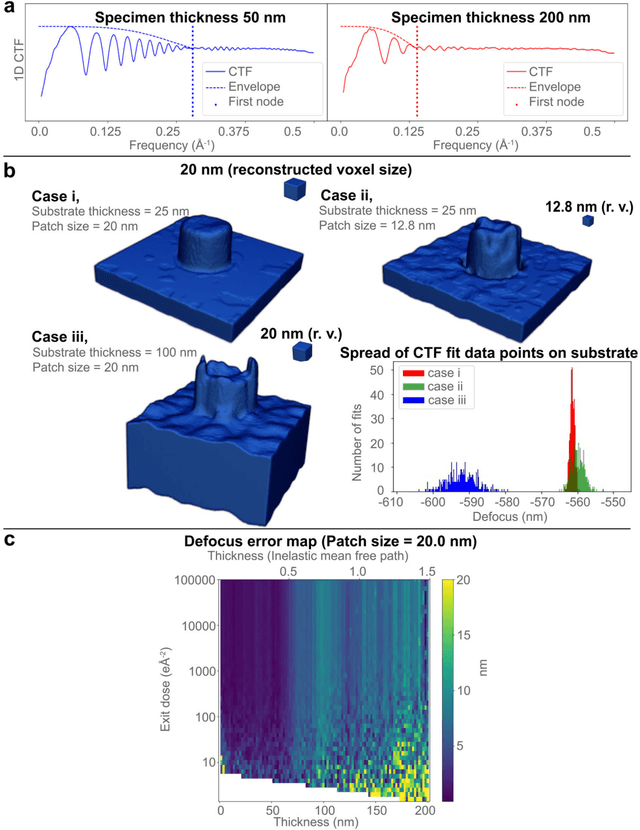
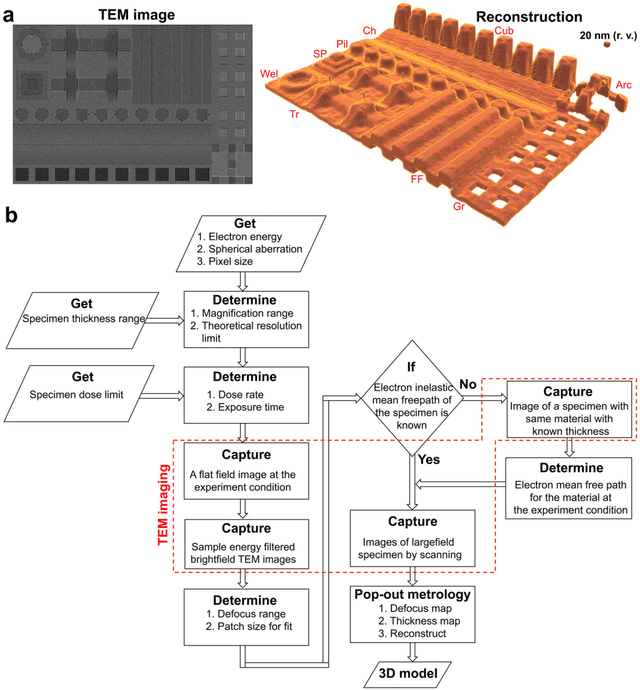
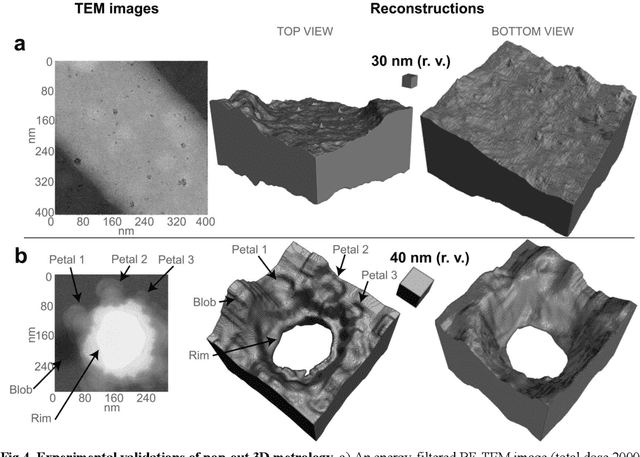
Abstract:Three-dimensional (3D) imaging of thin, extended specimens at nanometer resolution is critical for applications in biology, materials science, advanced synthesis, and manufacturing. Many 3D imaging techniques are limited to surface features, or available only for selective cross-sections, or require a tilt series of a local region, hence making them unsuitable for rapid, non-sacrificial screening of extended objects, or investigating fast dynamics. Here we describe a coherent imaging technique that recovers the 3D volume of a thin specimen with only a single, non-tomographic, energy-filtered, bright-field transmission electron microscopy (TEM) image. This technique does not require physically fracturing or sectioning thin specimens, only needs a single brief exposures to electron doses of ~100 e {\AA}-2, and can be readily calibrated for many existing TEMs; thus it can be widely deployed for rapid 3D metrology that complements existing forms of metrology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge