Virginie Uhlmann
Are Vision Foundation Models Foundational for Electron Microscopy Image Segmentation?
Feb 09, 2026Abstract:Although vision foundation models (VFMs) are increasingly reused for biomedical image analysis, it remains unclear whether the latent representations they provide are general enough to support effective transfer and reuse across heterogeneous microscopy image datasets. Here, we study this question for the problem of mitochondria segmentation in electron microscopy (EM) images, using two popular public EM datasets (Lucchi++ and VNC) and three recent representative VFMs (DINOv2, DINOv3, and OpenCLIP). We evaluate two practical model adaptation regimes: a frozen-backbone setting in which only a lightweight segmentation head is trained on top of the VFM, and parameter-efficient fine-tuning (PEFT) via Low-Rank Adaptation (LoRA) in which the VFM is fine-tuned in a targeted manner to a specific dataset. Across all backbones, we observe that training on a single EM dataset yields good segmentation performance (quantified as foreground Intersection-over-Union), and that LoRA consistently improves in-domain performance. In contrast, training on multiple EM datasets leads to severe performance degradation for all models considered, with only marginal gains from PEFT. Exploration of the latent representation space through various techniques (PCA, Fréchet Dinov2 distance, and linear probes) reveals a pronounced and persistent domain mismatch between the two considered EM datasets in spite of their visual similarity, which is consistent with the observed failure of paired training. These results suggest that, while VFMs can deliver competitive results for EM segmentation within a single domain under lightweight adaptation, current PEFT strategies are insufficient to obtain a single robust model across heterogeneous EM datasets without additional domain-alignment mechanisms.
ShapeEmbed: a self-supervised learning framework for 2D contour quantification
Jul 01, 2025Abstract:The shape of objects is an important source of visual information in a wide range of applications. One of the core challenges of shape quantification is to ensure that the extracted measurements remain invariant to transformations that preserve an object's intrinsic geometry, such as changing its size, orientation, and position in the image. In this work, we introduce ShapeEmbed, a self-supervised representation learning framework designed to encode the contour of objects in 2D images, represented as a Euclidean distance matrix, into a shape descriptor that is invariant to translation, scaling, rotation, reflection, and point indexing. Our approach overcomes the limitations of traditional shape descriptors while improving upon existing state-of-the-art autoencoder-based approaches. We demonstrate that the descriptors learned by our framework outperform their competitors in shape classification tasks on natural and biological images. We envision our approach to be of particular relevance to biological imaging applications.
Noise contrastive estimation with soft targets for conditional models
Apr 22, 2024Abstract:Soft targets combined with the cross-entropy loss have shown to improve generalization performance of deep neural networks on supervised classification tasks. The standard cross-entropy loss however assumes data to be categorically distributed, which may often not be the case in practice. In contrast, InfoNCE does not rely on such an explicit assumption but instead implicitly estimates the true conditional through negative sampling. Unfortunately, it cannot be combined with soft targets in its standard formulation, hindering its use in combination with sophisticated training strategies. In this paper, we address this limitation by proposing a principled loss function that is compatible with probabilistic targets. Our new soft target InfoNCE loss is conceptually simple, efficient to compute, and can be derived within the framework of noise contrastive estimation. Using a toy example, we demonstrate shortcomings of the categorical distribution assumption of cross-entropy, and discuss implications of sampling from soft distributions. We observe that soft target InfoNCE performs on par with strong soft target cross-entropy baselines and outperforms hard target NLL and InfoNCE losses on popular benchmarks, including ImageNet. Finally, we provide a simple implementation of our loss, geared towards supervised classification and fully compatible with deep classification model trained with cross-entropy.
MIFA: Metadata, Incentives, Formats, and Accessibility guidelines to improve the reuse of AI datasets for bioimage analysis
Nov 22, 2023



Abstract:Artificial Intelligence methods are powerful tools for biological image analysis and processing. High-quality annotated images are key to training and developing new methods, but access to such data is often hindered by the lack of standards for sharing datasets. We brought together community experts in a workshop to develop guidelines to improve the reuse of bioimages and annotations for AI applications. These include standards on data formats, metadata, data presentation and sharing, and incentives to generate new datasets. We are positive that the MIFA (Metadata, Incentives, Formats, and Accessibility) recommendations will accelerate the development of AI tools for bioimage analysis by facilitating access to high quality training data.
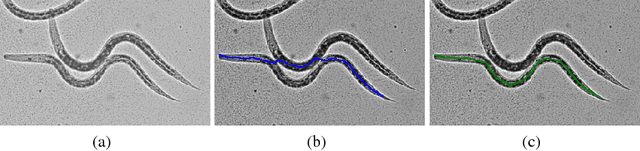
aura-net : robust segmentation of phase-contrast microscopy images with few annotations
Feb 02, 2021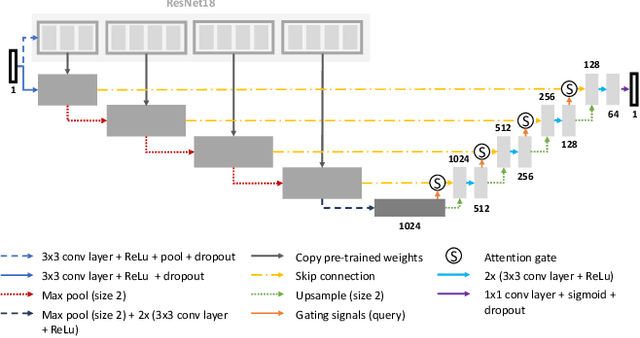
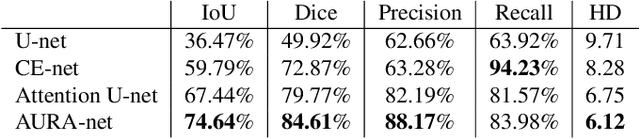
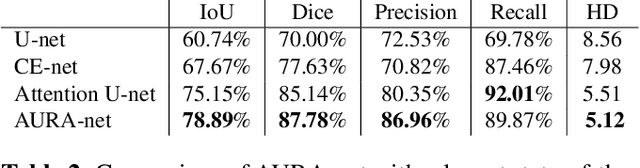
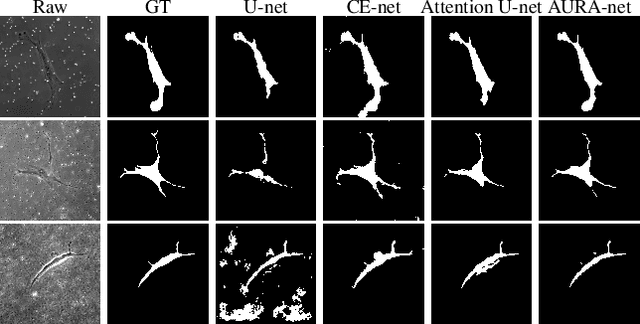
Abstract:We present AURA-net, a convolutional neural network (CNN) for the segmentation of phase-contrast microscopy images. AURA-net uses transfer learning to accelerate training and Attention mechanisms to help the network focus on relevant image features. In this way, it can be trained efficiently with a very limited amount of annotations. Our network can thus be used to automate the segmentation of datasets that are generally considered too small for deep learning techniques. AURA-net also uses a loss inspired by active contours that is well-adapted to the specificity of phase-contrast images, further improving performance. We show that AURA-net outperforms state-of-the-art alternatives in several small (less than 100images) datasets.
Diverse M-Best Solutions by Dynamic Programming
Mar 15, 2018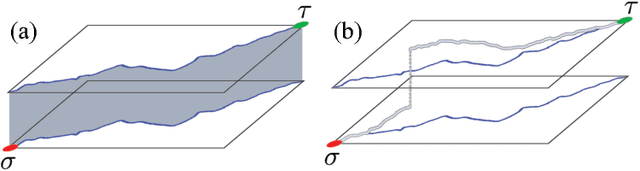
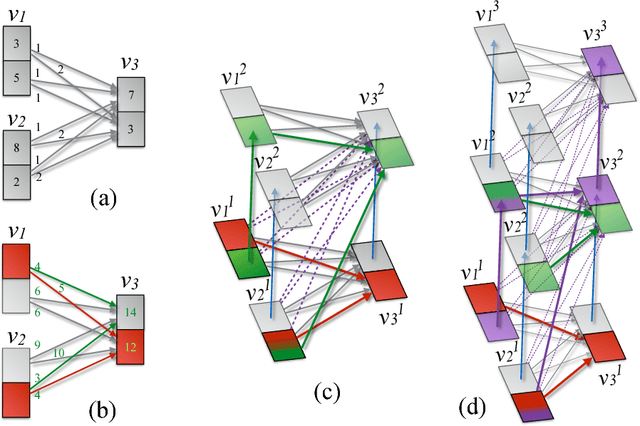
Abstract:Many computer vision pipelines involve dynamic programming primitives such as finding a shortest path or the minimum energy solution in a tree-shaped probabilistic graphical model. In such cases, extracting not merely the best, but the set of M-best solutions is useful to generate a rich collection of candidate proposals that can be used in downstream processing. In this work, we show how M-best solutions of tree-shaped graphical models can be obtained by dynamic programming on a special graph with M layers. The proposed multi-layer concept is optimal for searching M-best solutions, and so flexible that it can also approximate M-best diverse solutions. We illustrate the usefulness with applications to object detection, panorama stitching and centerline extraction. Note: We have observed that an assumption in section 4 of our paper is not always fulfilled, see the attached corrigendum for details.
* Includes supplementary and corrigendum
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge